Related Research Articles
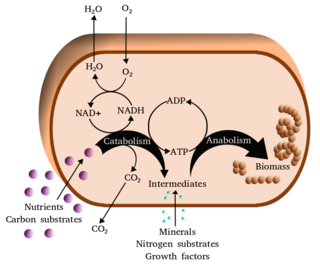
Metabolism is the set of life-sustaining chemical reactions in organisms. The three main functions of metabolism are: the conversion of the energy in food to energy available to run cellular processes; the conversion of food to building blocks of proteins, lipids, nucleic acids, and some carbohydrates; and the elimination of metabolic wastes. These enzyme-catalyzed reactions allow organisms to grow and reproduce, maintain their structures, and respond to their environments. The word metabolism can also refer to the sum of all chemical reactions that occur in living organisms, including digestion and the transportation of substances into and between different cells, in which case the above described set of reactions within the cells is called intermediary metabolism.
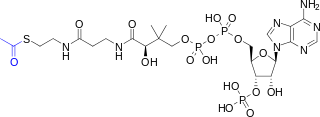
Acetyl-CoA is a molecule that participates in many biochemical reactions in protein, carbohydrate and lipid metabolism. Its main function is to deliver the acetyl group to the citric acid cycle to be oxidized for energy production.

Systems biology is the computational and mathematical analysis and modeling of complex biological systems. It is a biology-based interdisciplinary field of study that focuses on complex interactions within biological systems, using a holistic approach to biological research.

Metabolic engineering is the practice of optimizing genetic and regulatory processes within cells to increase the cell's production of a certain substance. These processes are chemical networks that use a series of biochemical reactions and enzymes that allow cells to convert raw materials into molecules necessary for the cell's survival. Metabolic engineering specifically seeks to mathematically model these networks, calculate a yield of useful products, and pin point parts of the network that constrain the production of these products. Genetic engineering techniques can then be used to modify the network in order to relieve these constraints. Once again this modified network can be modeled to calculate the new product yield.

Henrik Kacser FRSE was a Austro-Hungarian-born biochemist and geneticist who worked in Britain in the 20th century. Kacser's achievements have been recognised by his election to the Royal Society of Edinburgh in 1990, by an honorary doctorate of the University of Bordeaux II in 1993.
Metabolic network modelling, also known as metabolic network reconstruction or metabolic pathway analysis, allows for an in-depth insight into the molecular mechanisms of a particular organism. In particular, these models correlate the genome with molecular physiology. A reconstruction breaks down metabolic pathways into their respective reactions and enzymes, and analyzes them within the perspective of the entire network. In simplified terms, a reconstruction collects all of the relevant metabolic information of an organism and compiles it in a mathematical model. Validation and analysis of reconstructions can allow identification of key features of metabolism such as growth yield, resource distribution, network robustness, and gene essentiality. This knowledge can then be applied to create novel biotechnology.
Paracoccus denitrificans, is a coccoid bacterium known for its nitrate reducing properties, its ability to replicate under conditions of hypergravity and for being a relative of the eukaryotic mitochondrion.

Metabolic flux analysis (MFA) is an experimental fluxomics technique used to examine production and consumption rates of metabolites in a biological system. At an intracellular level, it allows for the quantification of metabolic fluxes, thereby elucidating the central metabolism of the cell. Various methods of MFA, including isotopically stationary metabolic flux analysis, isotopically non-stationary metabolic flux analysis, and thermodynamics-based metabolic flux analysis, can be coupled with stoichiometric models of metabolism and mass spectrometry methods with isotopic mass resolution to elucidate the transfer of moieties containing isotopic tracers from one metabolite into another and derive information about the metabolic network. Metabolic flux analysis (MFA) has many applications such as determining the limits on the ability of a biological system to produce a biochemical such as ethanol, predicting the response to gene knockout, and guiding the identification of bottleneck enzymes in metabolic networks for metabolic engineering efforts.
Fluxomics describes the various approaches that seek to determine the rates of metabolic reactions within a biological entity. While metabolomics can provide instantaneous information on the metabolites in a biological sample, metabolism is a dynamic process. The significance of fluxomics is that metabolic fluxes determine the cellular phenotype. It has the added advantage of being based on the metabolome which has fewer components than the genome or proteome.
Moiety conservation is the conservation of a subgroup in a chemical species, which is cyclically transferred from one molecule to another. In biochemistry, moiety conservation can have profound effects on the system's dynamics.

Reinhart Heinrich was a German biophysicist.

Pyruvate dehydrogenase kinase isoform 2 (PDK2) also known as pyruvate dehydrogenase lipoamide kinase isozyme 2, mitochondrial is an enzyme that in humans is encoded by the PDK2 gene. PDK2 is an isozyme of pyruvate dehydrogenase kinase.

Douglas Bruce Kell is a British biochemist and Research Professor of Systems Biology in the Institute of Systems, Molecular and Integrative Biology at the University of Liverpool. He was previously at the School of Chemistry at the University of Manchester, based in the Manchester Institute of Biotechnology (MIB). He founded and led the Manchester Centre for Integrative Systems Biology. He served as chief executive officer (CEO) of the Biotechnology and Biological Sciences Research Council (BBSRC) from 2008 to 2013.

Jens Nielsen is the CEO of BioInnovation Institute, Copenhagen, Denmark, and professor of systems biology at Chalmers University of Technology, Gothenburg, Sweden. He is also an adjunct professor at the Technical University of Denmark. Nielsen is the most cited researcher in the field of metabolic engineering, and he is the founding president of the International Metabolic Engineering Society. He has additionally founded several biotech companies.

Stefan Schuster is a German biophysicist. He is professor for bioinformatics at the University of Jena.
Metabolite damage can occur through enzyme promiscuity or spontaneous chemical reactions. Many metabolites are chemically reactive and unstable and can react with other cell components or undergo unwanted modifications. Enzymatically or chemically damaged metabolites are always useless and often toxic. To prevent toxicity that can occur from the accumulation of damaged metabolites, organisms have damage-control systems that:
- Reconvert damaged metabolites to their original, undamaged form
- Convert a potentially harmful metabolite to a benign one
- Prevent damage from happening by limiting the build-up of reactive, but non-damaged metabolites that can lead to harmful products
Robert L. Last is a plant biochemical genomicist who studies metabolic processes that protect plants from the environment and produce products important for animal and human nutrition. His research has covered (1) production and breakdown of essential amino acids, (2) the synthesis and protective roles of Vitamin C and Vitamin E (tocopherols) as well as identification of mechanisms that protect photosystem II from damage, and (3) synthesis and biological functions of plant protective specialized metabolites. Four central questions are: (i) how are leaf and seed amino acids levels regulated, (ii.) what mechanisms protect and repair photosystem II from stress-induced damage, (iii.) how do plants produce protective metabolites in their glandular secreting trichomes (iv.) and what are the evolutionary mechanisms that contribute to the tremendous diversity of specialized metabolites that protect plants from insects and pathogens and are used as therapeutic agents.
The Portland Press Excellence in Science Award was an annual award instituted in 1964 to recognize notable research in any branch of biochemistry undertaken in the UK or Republic of Ireland. It was initially called the CIBA Medal and Prize, then the Novartis Medal and Prize. The prize consists of a medal and a £3000 cash award. The winner is invited to present a lecture at a Society conference and submit an article to one of the Society's publications. Notable recipients include the Nobel laureates John E. Walker, Paul Nurse, Sydney Brenner, César Milstein, Peter D. Mitchell, Rodney Porter, and John Cornforth.
In biochemistry, a rate-limiting step is a step that controls the rate of a series of biochemical reactions. The statement is, however, a misunderstanding of how a sequence of enzyme catalyzed reaction steps operate. Rather than a single step controlling the rate, it has been discovered that multiple steps control the rate. Moreover, each controlling step controls the rate to varying degrees.
In metabolic control analysis, a variety of theorems have been discovered and discussed in the literature. The most well known of these are flux and concentration control coefficient summation relationships. These theorems are the result of the stoichiometric structure and mass conservation properties of biochemical networks. Equivalent theorems have not been found, for example, in electrical or economic systems.
References
- 1 2 "Professor David Fell - Professor of Systems Biology". Oxford Brookes University.
- ↑ "David Fell - Google Scholar Citations". scholar.google.no. Retrieved 31 August 2020.
- 1 2 Fell, David (1997). Understanding the control of metabolism . Portland Press. ISBN 9781855780477. OCLC 553392040.
- ↑ Fell, David A.; Liddle, Peter F.; Peacocke, Arthur R.; Dwek, Raymond A. (1974). "The preparation and properties of pyruvate kinase from yeast". Biochemical Journal. 139 (3): 665–675. doi:10.1042/bj1390665. ISSN 0264-6021. PMC 1166331 . PMID 4369339.
- ↑ Fell, David (1978). "An automated mixing apparatus for determining haemoglobin-oxygen dissociation". The Journal of Physiology. 2823: 3P–4P. doi:10.1113/jphysiol.1978.sp012483. PMID 31462. S2CID 222217462.
- ↑ El-Yassin, D I; Fell, D A; Lloyd, B B; Fisher, R B (1979). "The breakdown of 2,3-bis(phospho)-D-glycerate by Fe(III)-haemoglobin". Biochemical Journal. 177 (1): 373–375. doi:10.1042/bj1770373. ISSN 0264-6021. PMC 1186379 . PMID 426777.
- ↑ Fell, David A. (1980). "Theoretical analyses of the functioning of the high- and low-Km cyclic nucleotide phosphodiesterases in the regulation of the concentration of adenosine 3′,5′-cyclic monophosphate in animal cells". Journal of Theoretical Biology. 84 (2): 361–385. doi:10.1016/s0022-5193(80)80011-7. ISSN 0022-5193. PMID 6251314.
- ↑ Fell, D A; Sauro, H M (1986). "Metabolic control and its analysis: additional relationships between elasticities and control coefficients". European Journal of Biochemistry. 148 (3): 555–5616. doi: 10.1111/j.1432-1033.1985.tb08876.x . ISSN 0014-2956. PMID 3996393.
- ↑ Fell, D A; Small, J R (1986). "Fat synthesis in adipose tissue. An examination of stoichiometric constraints". Biochemical Journal. 238 (3): 781–786. doi:10.1042/bj2380781. ISSN 0264-6021. PMC 1147204 . PMID 3800960.
- ↑ Moniz-Barreto, P.; Fell, D. A. (1996), "Simulation of Dioxygen Free Radical Reactions: Their Importance in the Initiation of Lipid Peroxidation", Biomedical and Life Physics, Vieweg+Teubner Verlag, pp. 137–144, doi:10.1007/978-3-322-85017-1_12, ISBN 9783322850195
- ↑ Schuster, S.; Hilgetag, C.; Schuster, R. (1996), "Determining Elementary Modes of Functioning in Biochemical Reaction Networks at Steady State", Biomedical and Life Physics, Vieweg+Teubner Verlag, pp. 101–114, doi:10.1007/978-3-322-85017-1_9, ISBN 9783322850195
- ↑ Poolman, Mark G.; Fell, David A.; Raines, Christine A. (2003). "Elementary modes analysis of photosynthate metabolism in the chloroplast stroma". European Journal of Biochemistry. 270 (3): 430–439. doi: 10.1046/j.1432-1033.2003.03390.x . ISSN 0014-2956. PMID 12542693.
- ↑ Poolman, M. G.; Miguet, L.; Sweetlove, L. J.; Fell, D. A. (2009). "A Genome-Scale Metabolic Model of Arabidopsis and Some of Its Properties". Plant Physiology. 151 (3): 1570–1581. doi:10.1104/pp.109.141267. ISSN 0032-0889. PMC 2773075 . PMID 19755544.
- ↑ "David Fell - Google Scholar Citations". scholar.google.no. Retrieved 25 September 2018.
- ↑ Kacser, H; Burns, JA; Fell, DA (1995). "The control of flux". Biochemical Society Transactions. 23 (2): 341–366. doi:10.1042/bst0230341. PMID 7672373.
- ↑ Kacser, H; Burns, J A (1973). "The control of flux". Symposia of the Society for Experimental Biology. 27: 65–104.