Related Research Articles

Henrik Kacser FRSE was a Austro-Hungarian-born biochemist and geneticist who worked in Britain in the 20th century. Kacser's achievements have been recognised by his election to the Royal Society of Edinburgh in 1990, by an honorary doctorate of the University of Bordeaux II in 1993.

Hans Victor Westerhoff is a Dutch biologist and biochemist who is professor of synthetic systems biology at the University of Amsterdam and AstraZeneca professor of systems biology at the University of Manchester. Currently he is a Chair of AstraZeneca and a director of the Manchester Centre for Integrative Systems Biology.
The Systems Biology Markup Language (SBML) is a representation format, based on XML, for communicating and storing computational models of biological processes. It is a free and open standard with widespread software support and a community of users and developers. SBML can represent many different classes of biological phenomena, including metabolic networks, cell signaling pathways, regulatory networks, infectious diseases, and many others. It has been proposed as a standard for representing computational models in systems biology today.

The Systems Biology Ontology (SBO) is a set of controlled, relational vocabularies of terms commonly used in systems biology, and in particular in computational modeling.

Reinhart Heinrich was a German biophysicist.
Igor I. Goryanin is a systems biologist, who holds a Henrik Kacser Chair in Computational Systems Biology at the University of Edinburgh. He also heads the Biological Systems Unit at the Okinawa Institute of Science and Technology, Japan.
In enzymology, a 1-acylglycerol-3-phosphate O-acyltransferase is an enzyme that catalyzes the chemical reaction

In enzymology, a guanylate kinase is an enzyme that catalyzes the chemical reaction
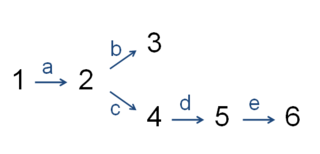
In biochemistry, the committed step is an effectively irreversible, enzyme-catalyzed reaction that occurs at a branch point during the biosynthesis of some molecules. As the name implies, after this step, the molecules are "committed" to the pathway and will ultimately end up in the pathway's final product. The first committed step should not be confused with the rate-limiting step, which is the step with the highest flux control coefficient. It is rare that the first committed step is in fact the rate-determining step.
LibSBML is an open-source software library that provides an application programming interface (API) for the SBML format. The libSBML library can be embedded in a software application or used in a web servlet as part of the application or servlet's implementation of support for reading, writing, and manipulating SBML documents and data streams. The core of libSBML is written in ISO standard C++; the library provides API for many programming languages via interfaces generated with the help of SWIG.

JSBML is an open-source Java (API) for the SBML format. Its API strives to attain a strong similarity to the Java binding of the corresponding library libSBML, but is entirely implemented in Java and therefore platform independent. JSBML provides an elaborated abstract type hierarchy, whose data types implement or extend many interfaces and abstract classes from the standard Java library. In this way, JSBML integrates smoothly into existing Java projects, and provides methods to read, write, evaluate, and manipulate the content of SBML documents.
Nicolas Le Novère is a British and French biologist. His research focuses on modeling signaling pathways and developing tools to share mathematical models.
David A. Fell is a British biochemist and professor of systems biology at Oxford Brookes University. He has published over 200 publications, including a textbook on Understanding the control of metabolism in 1996.

Athel Cornish-Bowden is a British biochemist known for his numerous textbooks, particularly those on enzyme kinetics and his work on metabolic control analysis.
Jan-Hendrik HofmeyrFRSSAf is one of the leaders in the field of metabolic control analysis and the quantitative analysis of metabolic regulation.
In biochemistry, a rate-limiting step is a reaction step that controls the rate of a series of biochemical reactions. The statement is, however, a misunderstanding of how a sequence of enzyme-catalyzed reaction steps operate. Rather than a single step controlling the rate, it has been discovered that multiple steps control the rate. Moreover, each controlling step controls the rate to varying degrees.
libRoadRunner is a C/C++ software library that supports simulation of SBML based models.. It uses LLVM to generate extremely high-performance code and is the fastest SBML-based simulator currently available. Its main purpose is for use as a reusable library that can be hosted by other applications, particularly on large compute clusters for doing parameter optimization where performance is critical. It also has a set of Python bindings that allow it to be easily used from Python as well as a set of bindings for Julia.
The classic Monod–Wyman–Changeux model (MWC) for cooperativity is generally published in an irreversible form. That is, there are no product terms in the rate equation which can be problematic for those wishing to build metabolic models since there are no product inhibition terms. However, a series of publications by Popova and Sel'kov derived the MWC rate equation for the reversible, multi-substrate, multi-product reaction.
In metabolic control analysis, a variety of theorems have been discovered and discussed in the literature. The most well known of these are flux and concentration control coefficient summation relationships. These theorems are the result of the stoichiometric structure and mass conservation properties of biochemical networks. Equivalent theorems have not been found, for example, in electrical or economic systems.
References
- ↑ ORCID 0000-0002-3659-6817
- ↑ Fell, D A; Sauro, H M (1985). "Metabolic control and its analysis: additional relationships between elasticities and control coefficients". Eur. J. Biochem. 148 (3): 555–561. doi: 10.1111/j.1432-1033.1985.tb08876.x . PMID 3996393.
- ↑ Acerenza, L; Sauro, H M; Kacser, H (1989). "Control analysis of time-dependent metabolic systems". J. Theor. Biol. 137 (4): 423–444. Bibcode:1989JThBi.137..423A. doi:10.1016/S0022-5193(89)80038-4. PMID 2626059.
- ↑ Kacser, H; Sauro, H M; Acerenza, L (1990). "Enzyme-enzyme interactions and control analysis. 1. the case of nonadditivity - monomer-oligomer associations". Eur. J. Biochem. 187 (3): 481–491. doi: 10.1111/j.1432-1033.1990.tb15329.x . PMID 2406132.
- ↑ Sauro, H M; Kacser, H (1990). "Enzyme-enzyme interactions and control analysis. 2. The case of nonindependence - heterologous association". Eur. J. Biochem. 187 (3): 493–500. doi: 10.1111/j.1432-1033.1990.tb15330.x . PMID 2406133.
- ↑ Sauro, H M (1993). "SCAMP: a general-purpose simulator and metabolic control analysis programs". Comput. Appl. Biosci. 9 (4): 441–450. doi:10.1093/bioinformatics/9.4.441. PMID 8402211.
- ↑ Sauro, H M (2000). "Jarnac: a system for interactive metabolic analysis". In Hofmeyr, J.-H.S.; Rohwer, J. M; Snoep, J. L (eds.). Animating the Cellular Map. Stellenbosch University Press. pp. 221–228. ISBN 0-7972-0776-7.
- ↑ Wellock, C; Chickarmane, V; Sauro, H M (2005). "The SBW-MATLAB Interface". Bioinformatics. 21 (6): 823–824. doi:10.1093/bioinformatics/bti110. PMID 15531613.
- ↑ Hucka, M.; Finney, A.; Sauro, H. M.; Bolouri, H.; Doyle, J. C.; Kitano, H.; Arkin, A. P.; Bornstein, B. J.; Bray, D; Cornish-Bowden, A.; Cuellar, A. A.; Dronov, S.; Gilles, E.D.; Ginkel, M; Gor, V.; Goryanin, I.I.; Hedley, W.J.; Hodgman, T. C.; Hofmeyr, J.-H.; Hunter, P. J.; Juty, N. S.; Kasberger, J. L.; Kremling, A.; Kummer, U.; Le Novère, N.; Loew, L. M.; Lucio, D.; Mendes, P.; Minch, E.; Mjolsness, E.D.; Nakayama, Y.; Nelson, M.R.; Nielsen, P. F.; Sakurada, T.; Schaff, J. C.; Shapiro, B.E.; Shimizu, T. S.; Spence, H. D.; Stelling, J.; Takahashi, K.; Tomita, M.; Wagner, J.; Wang, J. (2003). "The systems biology markup language (SBML): A medium for representation and exchange of biochemical network models". Bioinformatics. 19 (4): 524–531. CiteSeerX 10.1.1.562.1085 . doi: 10.1093/bioinformatics/btg015 . PMID 12611808.
- ↑ More than 3800 citations as of late 2024.
- ↑ Choi, Kiri; Medley, J. Kyle; König, Matthias; Stocking, Kaylene; Smith, Lucian; Gu, Stanley; Sauro, Herbert M. (July 2018). "Tellurium: An extensible python-based modeling environment for systems and synthetic biology". BioSystems. 171: 74–79. doi:10.1016/j.biosystems.2018.07.006. ISSN 1872-8324. PMC 6108935 . PMID 30053414.
- ↑ Sauro, H M (2018). Systems Biology: An Introduction to Metabolic Control Analysis (2nd ed.). Ambrosius. ISBN 978-0982477366.
- ↑ Sauro, H M (2023). Enzyme Kinetics for Systems Biology (2nd ed.). Ambrosius Publishing. ISBN 978-0982477335.
- ↑ Sauro, H M (2021). The Little Book of Laplace Transforms. Ambrosius Publishing. ISBN 978-1732548619.
- ↑ https://depts.washington.edu/bpsd/people/faculty/sauro/
- ↑ "Herbert M Sauro". ResearchGate. 14 March 2023. Retrieved 14 March 2023.
- ↑ "Past award recipients". UW College of Engineering. 25 June 2009. Retrieved 15 March 2023.
- ↑ "Center for Reproducible Biomedical Modeling". Center for Reproducible Biomedical Modeling.