
A protease is an enzyme that catalyzes proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in many biological functions, including digestion of ingested proteins, protein catabolism, and cell signaling.
In biology and biochemistry, protease inhibitors, or antiproteases, are molecules that inhibit the function of proteases. Many naturally occurring protease inhibitors are proteins.
A metalloproteinase, or metalloprotease, is any protease enzyme whose catalytic mechanism involves a metal. An example is ADAM12 which plays a significant role in the fusion of muscle cells during embryo development, in a process known as myogenesis.

A catalytic triad is a set of three coordinated amino acids that can be found in the active site of some enzymes. Catalytic triads are most commonly found in hydrolase and transferase enzymes. An acid-base-nucleophile triad is a common motif for generating a nucleophilic residue for covalent catalysis. The residues form a charge-relay network to polarise and activate the nucleophile, which attacks the substrate, forming a covalent intermediate which is then hydrolysed to release the product and regenerate free enzyme. The nucleophile is most commonly a serine or cysteine amino acid, but occasionally threonine or even selenocysteine. The 3D structure of the enzyme brings together the triad residues in a precise orientation, even though they may be far apart in the sequence.

Cathepsin C (CTSC) also known as dipeptidyl peptidase I (DPP-I) is a lysosomal exo-cysteine protease belonging to the peptidase C1 protein family, a subgroup of the cysteine cathepsins. In humans, it is encoded by the CTSC gene.
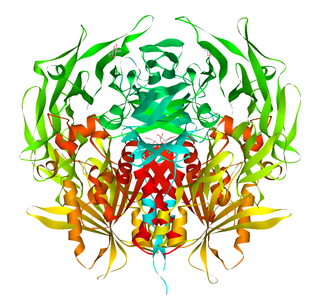
Dipeptidyl peptidase-4 (DPP4), also known as adenosine deaminase complexing protein 2 or CD26 is a protein that, in humans, is encoded by the DPP4 gene. DPP4 is related to FAP, DPP8, and DPP9. The enzyme was discovered in 1966 by Hopsu-Havu and Glenner, and as a result of various studies on chemism, was called dipeptidyl peptidase IV [DP IV].

Aspartic proteases are a catalytic type of protease enzymes that use an activated water molecule bound to one or more aspartate residues for catalysis of their peptide substrates. In general, they have two highly conserved aspartates in the active site and are optimally active at acidic pH. Nearly all known aspartyl proteases are inhibited by pepstatin.

Fibroblast activation protein alpha (FAP-alpha) also known as prolyl endopeptidase FAP is an enzyme that in humans is encoded by the FAP gene.

Dipeptidyl-peptidase 3 is an enzyme that in humans is encoded by the DPP3 gene.

Dipeptidyl aminopeptidase-like protein 6 is a protein that in humans is encoded by the DPP6 gene.

Inactive dipeptidyl peptidase 10 is a protein that in humans is encoded by the DPP10 gene. Alternate transcriptional splice variants, encoding different isoforms, have been characterized.

Dipeptidyl peptidase 9 is an enzyme that in humans is encoded by the DPP9 gene.
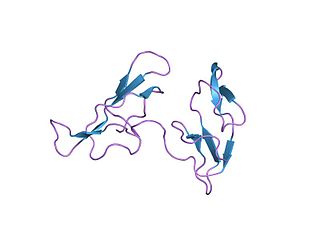
In molecular biology, the Bowman–Birk protease inhibitor family of proteins consists of eukaryotic proteinase inhibitors, belonging to MEROPS inhibitor family I12, clan IF. They mainly inhibit serine peptidases of the S1 family, but also inhibit S3 peptidases.
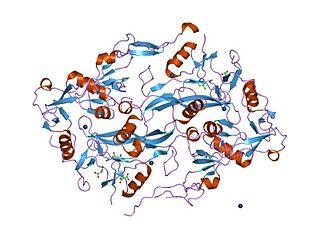
The Kazal domain is an evolutionary conserved protein domain usually indicative of serine protease inhibitors. However, kazal-like domains are also seen in the extracellular part of agrins, which are not known to be protease inhibitors.

In molecular biology, the CLP protease family is a family of serine peptidases belong to the MEROPS peptidase family S14. ClpP is an ATP-dependent protease that cleaves a number of proteins, such as casein and albumin. It exists as a heterodimer of ATP-binding regulatory A and catalytic P subunits, both of which are required for effective levels of protease activity in the presence of ATP, although the P subunit alone does possess some catalytic activity.

In molecular biology, the Lon protease family is a family of enzymes that break peptide bonds in proteins resulting in smaller peptides or amino acids. They are found in archaea, bacteria and eukaryotes. Lon proteases are ATP-dependent serine peptidases belonging to the MEROPS peptidase family S16. In the eukaryotes the majority of the Lon proteases are located in the mitochondrial matrix. In yeast, the Lon protease PIM1 is located in the mitochondrial matrix. It is required for mitochondrial function, it is constitutively expressed but is increased after thermal stress, suggesting that PIM1 may play a role in the heat shock response. Lon proteases have two specific subfamilies: LonA and LonB, differentiated by the number of AAA+ domains found in the protein.

The PA clan is the largest group of proteases with common ancestry as identified by structural homology. Members have a chymotrypsin-like fold and similar proteolysis mechanisms but can have identity of <10%. The clan contains both cysteine and serine proteases. PA clan proteases can be found in plants, animals, fungi, eubacteria, archaea and viruses.

Pacifastin is a family of serine proteinase inhibitors found in arthropods. Pacifastin inhibits the serine peptidases trypsin and chymotrypsin.
Asparagine peptide lyase are one of the seven groups in which proteases, also termed proteolytic enzymes, peptidases, or proteinases, are classified according to their catalytic residue. The catalytic mechanism of the asparagine peptide lyases involves an asparagine residue acting as nucleophile to perform a nucleophilic elimination reaction, rather than hydrolysis, to catalyse the breaking of a peptide bond.
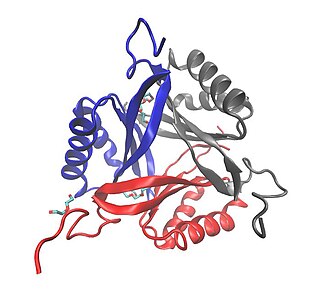
SbtB, which stands for sodium-bicarbonate-transporter B, is a protein found in bacteria. This small soluble protein has been classified as a new member of the P-II family that is involved in signal transduction. This protein has been demonstrated to participate in numerous processes including carbon sensing mechanisms in cyanobacteria.

















