In chemistry, a disulfide refers to a functional group with the structure R−S−S−R′. The linkage is also called an SS-bond or sometimes a disulfide bridge and is usually derived by the coupling of two thiol groups. In biology, disulfide bridges formed between thiol groups in two cysteine residues are an important component of the secondary and tertiary structure of proteins. The connection is a persulfide, in analogy to its congener, peroxide (R−O−O−R′), but this terminology is rarely used, except in reference to hydrodisulfides.

Post-translational modification (PTM) refers to the covalent and generally enzymatic modification of proteins following protein biosynthesis. Proteins are synthesized by ribosomes translating mRNA into polypeptide chains, which may then undergo PTM to form the mature protein product. PTMs are important components in cell signaling, as for example when prohormones are converted to hormones.
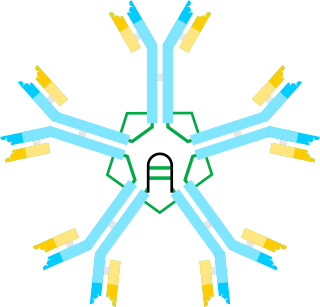
Immunoglobulin M (IgM) is one of several isotypes of antibody that are produced by vertebrates. IgM is the largest antibody, and it is the first antibody to appear in the response to initial exposure to an antigen. In the case of humans and other mammals that have been studied, the spleen, where plasmablasts responsible for antibody production reside, is the major site of specific IgM production.
Protein disulfide isomerase, or PDI, is an enzyme in the endoplasmic reticulum (ER) in eukaryotes and the periplasm of bacteria that catalyzes the formation and breakage of disulfide bonds between cysteine residues within proteins as they fold. This allows proteins to quickly find the correct arrangement of disulfide bonds in their fully folded state, and therefore the enzyme acts to catalyze protein folding.

Thioredoxin is a class of small redox proteins known to be present in all organisms. It plays a role in many important biological processes, including redox signaling. In humans, thioredoxins are encoded by TXN and TXN2 genes. Loss-of-function mutation of either of the two human thioredoxin genes is lethal at the four-cell stage of the developing embryo. Although not entirely understood, thioredoxin plays a central role in humans and is increasingly linked to medicine through their response to reactive oxygen species (ROS). In plants, thioredoxins regulate a spectrum of critical functions, ranging from photosynthesis to growth, flowering and the development and germination of seeds. They have also recently been found to play a role in cell-to-cell communication.
Oxidative protein folding is a process that is responsible for the formation of disulfide bonds between cysteine residues in proteins. The driving force behind this process is a redox reaction, in which electrons pass between several proteins and finally to a terminal electron acceptor.

Vitamin K epoxide reductase (VKOR) is an enzyme that reduces vitamin K after it has been oxidised in the carboxylation of glutamic acid residues in blood coagulation enzymes. VKOR is a member of a large family of predicted enzymes that are present in vertebrates, Drosophila, plants, bacteria and archaea. In some plant and bacterial homologues, the VKOR domain is fused with domains of the thioredoxin family of oxidoreductases.
The Cys-loop ligand-gated ion channel superfamily is composed of nicotinic acetylcholine, GABAA, GABAA-ρ, glycine, 5-HT3, and zinc-activated (ZAC) receptors that are composed of five protein subunits that form a pentameric arrangement around a central pore. There are usually 2 alpha subunits and 3 other beta, gamma, or delta subunits (some consist of 5 alpha subunits). The name "Cys-loop" is because this type of receptors possess a characteristic loop formed by 13 highly conserved amino acids between two cysteine (Cys) residues which form a disulfide bond, near the N-terminal extracellular domain.

ER oxidoreductin 1 (Ero1) is an oxidoreductase enzyme that catalyses the formation and isomerization of protein disulfide bonds in the endoplasmic reticulum (ER) of eukaryotes. ER Oxidoreductin 1 (Ero1) is a conserved, luminal, glycoprotein that is tightly associated with the ER membrane, and is essential for the oxidation of protein dithiols. Since disulfide bond formation is an oxidative process, the major pathway of its catalysis has evolved to utilise oxidoreductases, which become reduced during the thiol-disulfide exchange reactions that oxidise the cysteine thiol groups of nascent polypeptides. Ero1 is required for the introduction of oxidising equivalents into the ER and their direct transfer to protein disulfide isomerase (PDI), thereby ensuring the correct folding and assembly of proteins that contain disulfide bonds in their native state.
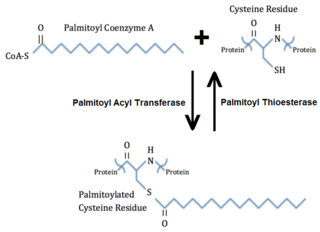
Palmitoylation is the covalent attachment of fatty acids, such as palmitic acid, to cysteine (S-palmitoylation) and less frequently to serine and threonine (O-palmitoylation) residues of proteins, which are typically membrane proteins. The precise function of palmitoylation depends on the particular protein being considered. Palmitoylation enhances the hydrophobicity of proteins and contributes to their membrane association. Palmitoylation also appears to play a significant role in subcellular trafficking of proteins between membrane compartments, as well as in modulating protein–protein interactions. In contrast to prenylation and myristoylation, palmitoylation is usually reversible. The reverse reaction in mammalian cells is catalyzed by acyl-protein thioesterases (APTs) in the cytosol and palmitoyl protein thioesterases in lysosomes. Because palmitoylation is a dynamic, post-translational process, it is believed to be employed by the cell to alter the subcellular localization, protein–protein interactions, or binding capacities of a protein.

An isopeptide bond is an amide bond that can form for example between the carboxyl group of one amino acid and the amino group of another. At least one of these joining groups is part of the side chain of one of these amino acids. This is unlike in a peptide bond which is sometimes called an eupeptide bond, especially when discussing about both of these bond types in the same context to make a distinction between the two.
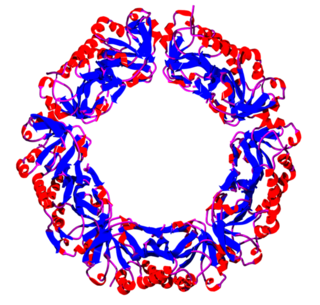
Peroxiredoxins are a ubiquitous family of antioxidant enzymes that also control cytokine-induced peroxide levels and thereby mediate signal transduction in mammalian cells. The family members in humans are PRDX1, PRDX2, PRDX3, PRDX4, PRDX5, and PRDX6. The physiological importance of peroxiredoxins is illustrated by their relative abundance. Peroxiredoxin Q is vitally important to plant chloroplasts, and by extension, plants to prevent excessive oxidative damage during stress, and yet unexplored role in human nutrition.

Protein disulfide-isomerase A3 (PDIA3), also known as glucose-regulated protein, 58-kD (GRP58), is an isomerase enzyme. This protein localizes to the endoplasmic reticulum (ER) and interacts with lectin chaperones calreticulin and calnexin (CNX) to modulate folding of newly synthesized glycoproteins. It is thought that complexes of lectins and this protein mediate protein folding by promoting formation of disulfide bonds in their glycoprotein substrates.

Discoidin domain is major protein domain of many blood coagulation factors.

The fimbrial usher protein is involved in biogenesis of the pilus in Gram-negative bacteria. The biogenesis of fimbriae requires a two-component assembly and transport system which is composed of a periplasmic chaperone and an outer membrane protein which has been termed a molecular 'usher'.

DsbA is a bacterial thiol disulfide oxidoreductase (TDOR). DsbA is a key component of the Dsb family of enzymes. DsbA catalyzes intrachain disulfide bond formation as peptides emerge into the cell's periplasm.
Somatomedin B is a serum factor of unknown function, is a small cysteine-rich peptide, derived proteolytically from the N-terminus of the cell-substrate adhesion protein vitronectin. Cys-rich somatomedin B-like domains are found in a number of proteins, including plasma-cell membrane glycoprotein and placental protein 11.

DsbC is a prokaryotic disulfide bond isomerase. The formation of native disulfide bonds play an important role in the proper folding of proteins and stabilize tertiary structures of the protein. DsbC is one of 6 proteins in the Dsb family in prokaryotes. The other proteins are DsbA, DsbB, DsbD, DsbE and DsbG. These enzymes work in tandem with each other to form disulfide bonds during the expression of proteins. DsbC and DsbG act as proofreaders of the disulfide bonds that are formed. They break non-native disulfide bonds that were formed and act as chaperones for the formation of native disulfide bonds. The isomerization of disulfide bonds occurs in the periplasm.

Bcl10-interacting CARD protein, also known as BinCARD, is a protein that in humans is encoded by the C9orf89 gene on chromosome 9. BinCARD is a member of the death-domain superfamily and contains a caspase recruitment domain (CARD). This protein regulates apoptosis and the immune response by inhibiting Bcl10, thus implicating it in diseases stemming from Bcl10 dysfunction.
The Disufide bond oxidoreductase D (DsbD) family is a member of the Lysine Exporter (LysE) Superfamily. A representative list of proteins belonging to the DsbD family can be found in the Transporter Classification Base.















