
Escherichia coli ( ESH-ə-RIK-ee-ə KOH-ly) is a gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus Escherichia that is commonly found in the lower intestine of warm-blooded organisms. Most E. coli strains are harmless, but some serotypes such as EPEC, and ETEC are pathogenic and can cause serious food poisoning in their hosts, and are occasionally responsible for food contamination incidents that prompt product recalls. Most strains are part of the normal microbiota of the gut and are harmless or even beneficial to humans (although these strains tend to be less studied than the pathogenic ones). For example, some strains of E. coli benefit their hosts by producing vitamin K2 or by preventing the colonization of the intestine by pathogenic bacteria. These mutually beneficial relationships between E. coli and humans are a type of mutualistic biological relationship — where both the humans and the E. coli are benefitting each other. E. coli is expelled into the environment within fecal matter. The bacterium grows massively in fresh fecal matter under aerobic conditions for three days, but its numbers decline slowly afterwards.

Escherichia coli O157:H7 is a serotype of the bacterial species Escherichia coli and is one of the Shiga-like toxin–producing types of E. coli. It is a cause of disease, typically foodborne illness, through consumption of contaminated and raw food, including raw milk and undercooked ground beef. Infection with this type of pathogenic bacteria may lead to hemorrhagic diarrhea, and to kidney failure; these have been reported to cause the deaths of children younger than five years of age, of elderly patients, and of patients whose immune systems are otherwise compromised.
The gene rpoS encodes the sigma factor sigma-38, a 37.8 kD protein in Escherichia coli. Sigma factors are proteins that regulate transcription in bacteria. Sigma factors can be activated in response to different environmental conditions. rpoS is transcribed in late exponential phase, and RpoS is the primary regulator of stationary phase genes. RpoS is a central regulator of the general stress response and operates in both a retroactive and a proactive manner: it not only allows the cell to survive environmental challenges, but it also prepares the cell for subsequent stresses (cross-protection). The transcriptional regulator CsgD is central to biofilm formation, controlling the expression of the curli structural and export proteins, and the diguanylate cyclase, adrA, which indirectly activates cellulose production. The rpoS gene most likely originated in the gammaproteobacteria.
A bacterial initiation factor (IF) is a protein that stabilizes the initiation complex for polypeptide translation.
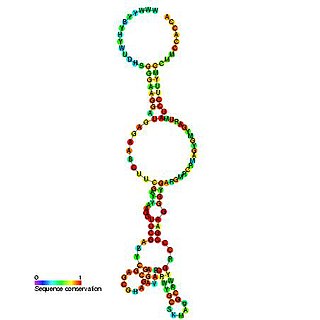
The RtT RNA is a RNA element that is released from the tyrT operon of Escherichia coli. The exact function of RtT is unknown although it is thought that it may be involved in changing the cellular response in relation to amino acid starvation.
Deoxyribonuclease IV (phage-T4-induced) is catalyzes the degradation nucleotides in DsDNA by attacking the 5'-terminal end.
Trimethylamine N-oxide reductase is a microbial enzyme that can reduce trimethylamine N-oxide (TMAO) into trimethylamine (TMA), as part of the electron transport chain. The enzyme has been purified from E. coli and the photosynthetic bacteria Roseobacter denitrificans.
In enzymology, a 4-phosphoerythronate dehydogenase (EC 1.1.1.290) is an enzyme that catalyzes the chemical reaction

In enzymology, a L-lysine 6-monooxygenase (NADPH) (EC 1.14.13.59) is an enzyme that catalyzes the chemical reaction
In enzymology, a furylfuramide isomerase is an enzyme that catalyzes the chemical reaction

The enzyme Acid-Induced Arginine Decarboxylase (AdiA), also commonly referred to as arginine decarboxylase, catalyzes the conversion of L-arginine into agmatine and carbon dioxide. The process consumes a proton in the decarboxylation and employs a pyridoxal-5'-phosphate (PLP) cofactor, similar to other enzymes involved in amino acid metabolism, such as ornithine decarboxylase and glutamine decarboxylase. It is found in bacteria and virus, though most research has so far focused on forms of the enzyme in bacteria. During the AdiA catalyzed decarboxylation of arginine, the necessary proton is consumed from the cell cytoplasm which helps to prevent the over-accumulation of protons inside the cell and serves to increase the intracellular pH. Arginine decarboxylase is part of an enzymatic system in Escherichia coli, Salmonella Typhimurium, and methane-producing bacteria Methanococcus jannaschii that makes these organisms acid resistant and allows them to survive under highly acidic medium.
In enzymology, a [acyl-carrier-protein] S-malonyltransferase is an enzyme that catalyzes the chemical reaction
4-Hydroxy-3-methylbut-2-enyl diphosphate reductase (EC 1.17.1.2, isopentenyl-diphosphate:NADP+ oxidoreductase, LytB, (E)-4-hydroxy-3-methylbut-2-en-1-yl diphosphate reductase, HMBPP reductase, IspH, LytB/IspH) is an enzyme in the non-mevalonate pathway. It acts upon (E)-4-Hydroxy-3-methyl-but-2-enyl pyrophosphate (or "HMB-PP").
Nitroreductases are a family of evolutionarily related proteins involved in the reduction of nitrogen-containing compounds, including those containing the nitro functional group. Members of this family utilise flavin mononucleotide as a cofactor and are often found to be homodimers.
Escherichia coli O104:H4 is an enteroaggregative Escherichia coli strain of the bacterium Escherichia coli, and the cause of the 2011 Escherichia coli O104:H4 outbreak. The "O" in the serological classification identifies the cell wall lipopolysaccharide antigen, and the "H" identifies the flagella antigen.
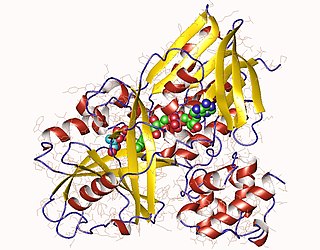
Glycerol-3-phosphate dehydrogenase (EC 1.1.5.3 is an enzyme with systematic name sn-glycerol 3-phosphate:quinone oxidoreductase. This enzyme catalyses the following chemical reaction
Formate dehydrogenase (acceptor) (EC 1.1.99.33, FDHH, FDH-H, FDH-O, formate dehydrogenase H, formate dehydrogenase O) is an enzyme with systematic name formate:acceptor oxidoreductase. This enzyme catalyses the following chemical reaction

Primary-amine oxidase, also known as semicarbazide-sensitive amine oxidase (SSAO), is an enzyme (EC 1.4.3.21) with the systematic name primary-amine:oxygen oxidoreductase (deaminating). This enzyme catalyses the following chemical reaction
Phosphoserine transaminase is an enzyme with systematic name O-phospho-L-serine:2-oxoglutarate aminotransferase. This enzyme catalyses the following chemical reaction
A. C. Matin is an Indian-American microbiologist, immunologist, academician and researcher. He is a professor of microbiology and immunology at Stanford University School of Medicine.






