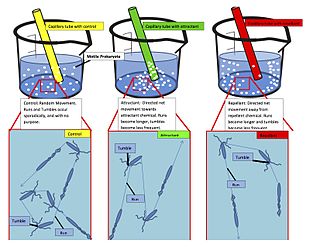
Chemotaxis is the movement of an organism or entity in response to a chemical stimulus. Somatic cells, bacteria, and other single-cell or multicellular organisms direct their movements according to certain chemicals in their environment. This is important for bacteria to find food by swimming toward the highest concentration of food molecules, or to flee from poisons. In multicellular organisms, chemotaxis is critical to early development and development as well as in normal function and health. In addition, it has been recognized that mechanisms that allow chemotaxis in animals can be subverted during cancer metastasis, and the aberrant change of the overall property of these networks, which control chemotaxis, can lead to carcinogenesis. The aberrant chemotaxis of leukocytes and lymphocytes also contribute to inflammatory diseases such as atherosclerosis, asthma, and arthritis. Sub-cellular components, such as the polarity patch generated by mating yeast, may also display chemotactic behavior.

A flagellum is a hair-like appendage that protrudes from certain plant and animal sperm cells, from fungal spores (zoospores), and from a wide range of microorganisms to provide motility. Many protists with flagella are known as flagellates.
dnaQ is the gene encoding the ε subunit of DNA polymerase III in Escherichia coli. The ε subunit is one of three core proteins in the DNA polymerase complex. It functions as a 3’→5’ DNA directed proofreading exonuclease that removes incorrectly incorporated bases during replication. dnaQ may also be referred to as mutD.
Flagellins are a family of proteins present in flagellated bacteria which arrange themselves in a hollow cylinder to form the filament in a bacterial flagellum. Flagellin has a mass on average of about 40,000 daltons. Flagellins are the principal component of bacterial flagella that have a crucial role in bacterial motility.

The L-ring of the bacterial flagellum is the ring in the lipid outer cell membrane through which the axial filament passes. that l ring stands for lipopolysaccharide.
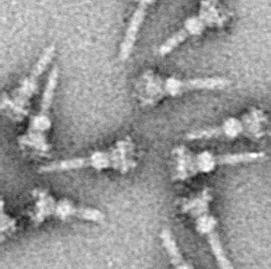
The type III secretion system is one of the bacterial secretion systems used by bacteria to secrete their effector proteins into the host's cells to promote virulence and colonisation. While the type III secretion system has been widely regarded as equivalent to the injectisome, many argue that the injectisome is only part of the type III secretion system, which also include structures like the flagellar export apparatus. The T3SS is a needle-like protein complex found in several species of pathogenic gram-negative bacteria.

In enzymology, a protein-glutamate O-methyltransferase is an enzyme that catalyzes the chemical reaction

The CorA transport system is the primary Mg2+ influx system of Salmonella typhimurium and Escherichia coli. CorA is ubiquitous in the Bacteria and Archaea. There are also eukaryotic members of the family localized to the mitochondrial membrane such as MRS2 and Lpe10 in yeast.

Bacterial motility is the ability of bacteria to move independently using metabolic energy. Most motility mechanisms that evolved among bacteria also evolved in parallel among the archaea. Most rod-shaped bacteria can move using their own power, which allows colonization of new environments and discovery of new resources for survival. Bacterial movement depends not only on the characteristics of the medium, but also on the use of different appendages to propel. Swarming and swimming movements are both powered by rotating flagella. Whereas swarming is a multicellular 2D movement over a surface and requires the presence of surfactants, swimming is movement of individual cells in liquid environments.
Motility protein A (MotA), is a bacterial protein that is encoded by the motA gene. It is a component of the flagellar motor. More specifically, MotA and MotB make the stator of a H+ driven bacterial flagella and surround the rotor as a ring of about 8–10 particles. MotA and MotB are integral membrane proteins. MotA has four transmembrane domains.
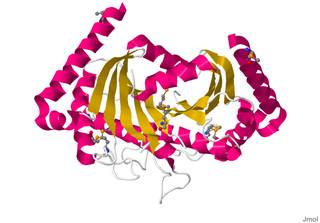
Motility protein B also known as MotB is a bacterial protein that is encoded by the motB gene. It's a component of the flagellar motor. More specifically, MotA and MotB makes the stator of a flagellum and surround the rotor as a ring of about 8-10 particles. MotA and MotB are integral membrane proteins. While both MotA and MotB surround the MS ring, MotB also anchors MotA to cell wall peptidoglycan. These two proteins form pores that harvest energy for flagellar mechanical movement by proton motive force (PMF) across the membrane. Cellular metabolic processes such as the electron transport chain move protons outside the cell, creating more protons and more positive charge in the extracellular space. When the protons flow back into the cell through MotA and MotB along concentration and charge gradients, they release energy that is used for flagellar rotation. The speed of the flagellar motor is dependent on the magnitude of the PMF acting on MotA and MotB.
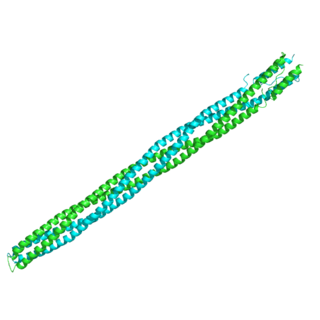
The methyl-accepting chemotaxis proteins are a family of transmembrane receptors that mediate chemotactic response in certain enteric bacteria, such as Salmonella enterica enterica and Escherichia coli. These methyl-accepting chemotaxis receptors are one of the first components in the sensory excitation and adaptation responses in bacteria, which act to alter swimming behaviour upon detection of specific chemicals. Use of the MCP allows bacteria to detect concentrations of molecules in the extracellular matrix so that the bacteria may smooth swim or tumble accordingly. If the bacterium detects rising levels of attractants (nutrients) or declining levels of repellents (toxins), the bacterium will continue swimming forward, or smooth swimming. If the bacterium detects declining levels of attractants or rising levels of repellents, the bacterium will tumble and re-orient itself in a new direction. In this manner, a bacterium may swim towards nutrients and away from toxins
The PilZ protein family is named after the type IV pilus control protein first identified in Pseudomonas aeruginosa, expressed as part of the pil operon. It has a cytoplasmic location and is essential for type IV fimbrial, or pilus, biogenesis. PilZ is a c-di-GMP binding domain and PilZ domain-containing proteins represent the best studied class of c-di-GMP effectors. C-di-GMP, cyclic diguanosine monophosphate, the second messenger in cells, is widespread in and unique to the bacterial kingdom. Elevated intracellular levels of c-di-GMP generally cause bacteria to change from a motile single-cell state to a sessile, adhesive surface-attached multicellular state called biofilm.
In molecular biology, bacterial DNA binding proteins are a family of small, usually basic proteins of about 90 residues that bind DNA and are known as histone-like proteins. Since bacterial binding proteins have a diversity of functions, it has been difficult to develop a common function for all of them. They are commonly referred to as histone-like and have many similar traits with the eukaryotic histone proteins. Eukaryotic histones package DNA to help it to fit in the nucleus, and they are known to be the most conserved proteins in nature. Examples include the HU protein in Escherichia coli, a dimer of closely related alpha and beta chains and in other bacteria can be a dimer of identical chains. HU-type proteins have been found in a variety of bacteria and archaea, and are also encoded in the chloroplast genome of some algae. The integration host factor (IHF), a dimer of closely related chains which is suggested to function in genetic recombination as well as in translational and transcriptional control is found in Enterobacteria and viral proteins including the African swine fever virus protein A104R.
In molecular biology, glycoside hydrolase family 73 is a family of glycoside hydrolases.
Histone-like nucleoid-structuring protein (H-NS), is one of twelve nucleoid-associated proteins (NAPs) whose main function is the organization of genetic material, including the regulation of gene expression via xenogeneic silencing. H-NS is characterized by an N-terminal domain (NTD) consisting of two dimerization sites, a linker region that is unstructured and a C-terminal domain (CTD) that is responsible for DNA-binding. Though it is a small protein, it provides essential nucleoid compaction and regulation of genes and is highly expressed, functioning as a dimer or multimer. Change in temperature causes H-NS to be dissociated from the DNA duplex, allowing for transcription by RNA polymerase, and in specific regions lead to pathogenic cascades in enterobacteria such as Escherichia coli and the four Shigella species.

In molecular biology, a response regulator is a protein that mediates a cell's response to changes in its environment as part of a two-component regulatory system. Response regulators are coupled to specific histidine kinases which serve as sensors of environmental changes. Response regulators and histidine kinases are two of the most common gene families in bacteria, where two-component signaling systems are very common; they also appear much more rarely in the genomes of some archaea, yeasts, filamentous fungi, and plants. Two-component systems are not found in metazoans.
The archaellum is a unique structure on the cell surface of many archaea that allows for swimming motility. The archaellum consists of a rigid helical filament that is attached to the cell membrane by a molecular motor. This molecular motor – composed of cytosolic, membrane, and pseudo-periplasmic proteins – is responsible for the assembly of the filament and, once assembled, for its rotation. The rotation of the filament propels archaeal cells in liquid medium, in a manner similar to the propeller of a boat. The bacterial analog of the archaellum is the flagellum, which is also responsible for their swimming motility and can also be compared to a rotating corkscrew. Although the movement of archaella and flagella is sometimes described as "whip-like", this is incorrect, as only cilia from Eukaryotes move in this manner. Indeed, even "flagellum" is a misnomer, as bacterial flagella also work as propeller-like structures.
CsgD is a transcription and response regulator protein referenced to as the master modulator of bacterial biofilm development. In E. coli cells, CsgD is tasked with aiding the transition from planktonic cell motility to the stationary phase of biofilm formation, in response to environmental growth factors. A transcription analysis assay illustrated a heightened decrease in CsgD's DNA-binding capacity when phosphorylated at A.A. D59 of the protein's primary sequence. Therefore, in the protein's active form (unphosphorylated), CsgD is capable of carrying out its normal functions of regulating curli proteins (fimbria) and producing ECM polysaccharides (cellulose). Following a promoter-lacZ fusion assay of CsgD binding to specific target sites on E. coli's genome, two classes of binding targets were identified: group I genes and group II genes. The group I genes, akin to fliE and yhbT, exhibit repressed transcription following their interaction with CsgD, whilst group II genes, including yccT and adrA, illustrated active functionality. Other group I operons that illustrate repressed transcription include fliE and fliEFGH, for motile flagellum formation. Other group II genes, imperative to the transition towards stationary biofilm development, include csgBA, encoding for curli fimbriae, and adrA, encoding for the synthesis of cyclic diguanylate. In this context, c-di-GMP functions as a bacterial secondary messenger, enhancing the production of extracellular cellulose and impeding flagellum production and rotation.
Michael Eisenbach is an Israeli biochemist who specializes in the navigation mechanisms of bacterial and sperm cells. He is a professor emeritus at the Weizmann Institute of Science, Department of Biomolecular Sciences, Rehovot, Israel. He discovered that sperm cells (spermatozoa) of mammals are actively guided to the egg. This opened the research field of mammalian sperm navigation. He demonstrated that the active navigation entails chemotaxis and thermotaxis. He made seminal contributions to the understanding of these two processes at the molecular, physiological and behavioural levels, as well as contributing to our understanding of the molecular mechanism of bacterial chemotaxis.













