
Evolution is the change in the heritable characteristics of biological populations over successive generations. It occurs when evolutionary processes such as natural selection and genetic drift act on genetic variation, resulting in certain characteristics becoming more or less common within a population over successive generations. The process of evolution has given rise to biodiversity at every level of biological organisation.

Sexual selection is a mechanism of evolution in which members of one biological sex choose mates of the other sex to mate with, and compete with members of the same sex for access to members of the opposite sex. These two forms of selection mean that some individuals have greater reproductive success than others within a population, for example because they are more attractive or prefer more attractive partners to produce offspring. Successful males benefit from frequent mating and monopolizing access to one or more fertile females. Females can maximise the return on the energy they invest in reproduction by selecting and mating with the best males.

Parasitism is a close relationship between species, where one organism, the parasite, lives on or inside another organism, the host, causing it some harm, and is adapted structurally to this way of life. The entomologist E. O. Wilson characterised parasites as "predators that eat prey in units of less than one". Parasites include single-celled protozoans such as the agents of malaria, sleeping sickness, and amoebic dysentery; animals such as hookworms, lice, mosquitoes, and vampire bats; fungi such as honey fungus and the agents of ringworm; and plants such as mistletoe, dodder, and the broomrapes.
Selfish genetic elements are genetic segments that can enhance their own transmission at the expense of other genes in the genome, even if this has no positive or a net negative effect on organismal fitness. Genomes have traditionally been viewed as cohesive units, with genes acting together to improve the fitness of the organism.

In biology, coevolution occurs when two or more species reciprocally affect each other's evolution through the process of natural selection. The term sometimes is used for two traits in the same species affecting each other's evolution, as well as gene-culture coevolution.
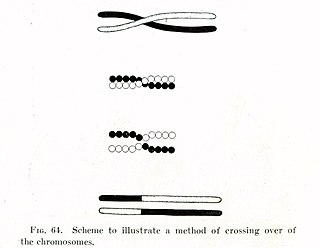
In evolutionary genetics, Muller's ratchet is a process which, in the absence of recombination, results in an accumulation of irreversible deleterious mutations. This happens because in the absence of recombination, and assuming reverse mutations are rare, offspring bear at least as much mutational load as their parents. Muller proposed this mechanism as one reason why sexual reproduction may be favored over asexual reproduction, as sexual organisms benefit from recombination and consequent elimination of deleterious mutations. The negative effect of accumulating irreversible deleterious mutations may not be prevalent in organisms which, while they reproduce asexually, also undergo other forms of recombination. This effect has also been observed in those regions of the genomes of sexual organisms that do not undergo recombination.

Genetic diversity is the total number of genetic characteristics in the genetic makeup of a species. It ranges widely, from the number of species to differences within species, and can be correlated to the span of survival for a species. It is distinguished from genetic variability, which describes the tendency of genetic characteristics to vary.

In biology, polymorphism is the occurrence of two or more clearly different morphs or forms, also referred to as alternative phenotypes, in the population of a species. To be classified as such, morphs must occupy the same habitat at the same time and belong to a panmictic population.

In population genetics, directional selection is a type of natural selection in which one extreme phenotype is favored over both the other extreme and moderate phenotypes. This genetic selection causes the allele frequency to shift toward the chosen extreme over time as allele ratios change from generation to generation. The advantageous extreme allele will increase as a consequence of survival and reproduction differences among the different present phenotypes in the population. The allele fluctuations as a result of directional selection can be independent of the dominance of the allele, and in some cases if the allele is recessive, it can eventually become fixed in the population.

Evolution of sexual reproduction describes how sexually reproducing animals, plants, fungi and protists could have evolved from a common ancestor that was a single-celled eukaryotic species. Sexual reproduction is widespread in eukaryotes, though a few eukaryotic species have secondarily lost the ability to reproduce sexually, such as Bdelloidea, and some plants and animals routinely reproduce asexually without entirely having lost sex. The evolution of sexual reproduction contains two related yet distinct themes: its origin and its maintenance. Bacteria and Archaea (prokaryotes) have processes that can transfer DNA from one cell to another, but it is unclear if these processes are evolutionarily related to sexual reproduction in Eukaryotes. In eukaryotes, true sexual reproduction by meiosis and cell fusion is thought to have arisen in the last eukaryotic common ancestor, possibly via several processes of varying success, and then to have persisted.
Evolutionary game theory (EGT) is the application of game theory to evolving populations in biology. It defines a framework of contests, strategies, and analytics into which Darwinian competition can be modelled. It originated in 1973 with John Maynard Smith and George R. Price's formalisation of contests, analysed as strategies, and the mathematical criteria that can be used to predict the results of competing strategies.

The viviparous lizard, or common lizard, is a Eurasian lizard. It lives farther north than any other species of non-marine reptile, and is named for the fact that it is viviparous, meaning it gives birth to live young. Both "Zootoca" and "vivipara" mean "live birth", in (Latinized) Greek and Latin respectively. It was called Lacerta vivipara until the genus Lacerta was split into nine genera in 2007 by Arnold, Arribas & Carranza.

Mate choice is one of the primary mechanisms under which evolution can occur. It is characterized by a "selective response by animals to particular stimuli" which can be observed as behavior. In other words, before an animal engages with a potential mate, they first evaluate various aspects of that mate which are indicative of quality—such as the resources or phenotypes they have—and evaluate whether or not those particular trait(s) are somehow beneficial to them. The evaluation will then incur a response of some sort.
The Red Queen's hypothesis is a hypothesis in evolutionary biology proposed in 1973, that species must constantly adapt, evolve, and proliferate in order to survive while pitted against ever-evolving opposing species. The hypothesis was intended to explain the constant (age-independent) extinction probability as observed in the paleontological record caused by co-evolution between competing species; however, it has also been suggested that the Red Queen hypothesis explains the advantage of sexual reproduction at the level of individuals, and the positive correlation between speciation and extinction rates in most higher taxa.
Host–parasite coevolution is a special case of coevolution, where a host and a parasite continually adapt to each other. This can create an evolutionary arms race between them. A more benign possibility is of an evolutionary trade-off between transmission and virulence in the parasite, as if it kills its host too quickly, the parasite will not be able to reproduce either. Another theory, the Red Queen hypothesis, proposes that since both host and parasite have to keep on evolving to keep up with each other, and since sexual reproduction continually creates new combinations of genes, parasitism favours sexual reproduction in the host.
Ecoimmunology or Ecological Immunology is the study of the causes and consequences of variation in immunity. The field of ecoimmunology seeks to give an ultimate perspective for proximate mechanisms of immunology. This approach places immunology in evolutionary and ecological contexts across all levels of biological organization.
Interlocus sexual conflict is a type of sexual conflict that occurs through the interaction of a set of antagonistic alleles at two or more different loci, or the location of a gene on a chromosome, in males and females, resulting in the deviation of either or both sexes from the fitness optima for the traits. A co-evolutionary arms race is established between the sexes in which either sex evolves a set of antagonistic adaptations that is detrimental to the fitness of the other sex. The potential for reproductive success in one organism is strengthened while the fitness of the opposite sex is weakened. Interlocus sexual conflict can arise due to aspects of male–female interactions such as mating frequency, fertilization, relative parental effort, female remating behavior, and female reproductive rate.

Major histocompatibility complex (MHC) genes code for cell surface proteins that facilitate an organism's immune response to pathogens as well as its ability to avoid attacking its own cells. These genes have maintained an unusually high level of allelic diversity throughout time and throughout different populations. This means that for each MHC gene, many alleles consistently exist within the population, and many individuals are heterozygous at MHC loci.

Evolving digital ecological networks are webs of interacting, self-replicating, and evolving computer programs that experience the same major ecological interactions as biological organisms. Despite being computational, these programs evolve quickly in an open-ended way, and starting from only one or two ancestral organisms, the formation of ecological networks can be observed in real-time by tracking interactions between the constantly evolving organism phenotypes. These phenotypes may be defined by combinations of logical computations that digital organisms perform and by expressed behaviors that have evolved. The types and outcomes of interactions between phenotypes are determined by task overlap for logic-defined phenotypes and by responses to encounters in the case of behavioral phenotypes. Biologists use these evolving networks to study active and fundamental topics within evolutionary ecology.
Disease ecology is a sub-discipline of ecology concerned with the mechanisms, patterns, and effects of host-pathogen interactions, particularly those of infectious diseases. For example, it examines how parasites spread through and influence wildlife populations and communities. By studying the flow of diseases within the natural environment, scientists seek to better understand how changes within our environment can shape how pathogens, and other diseases, travel. Therefore, diseases ecology seeks to understand the links between ecological interactions and disease evolution. New emerging and re-emerging infectious diseases are increasing at unprecedented rates which can have lasting impacts on public health, ecosystem health, and biodiversity.













