
Yersinia pestis is a gram-negative, non-motile, coccobacillus bacterium without spores that is related to both Yersinia pseudotuberculosis and Yersinia enterocolitica. It is a facultative anaerobic organism that can infect humans via the Oriental rat flea. It causes the disease plague, which caused the first plague pandemic and the Black Death, the deadliest pandemic in recorded history. Plague takes three main forms: pneumonic, septicemic, and bubonic.

Escherichia coli, also known as E. coli, is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus Escherichia that is commonly found in the lower intestine of warm-blooded organisms. Most E. coli strains are harmless, but some serotypes (EPEC, ETEC etc.) can cause serious food poisoning in their hosts, and are occasionally responsible for food contamination incidents that prompt product recalls. The harmless strains are part of the normal microbiota of the gut, and can benefit their hosts by producing vitamin K2, and preventing colonisation of the intestine with pathogenic bacteria, having a mutualistic relationship. E. coli is expelled into the environment within fecal matter. The bacterium grows massively in fresh fecal matter under aerobic conditions for three days, but its numbers decline slowly afterwards.
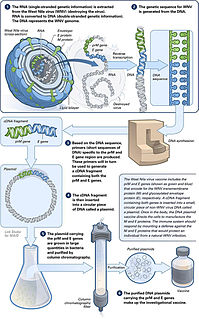
A DNA vaccine is a type of vaccine that transfects a specific antigen-coding DNA sequence into the cells of an organism as a mechanism to induce an immune response.

Treponema pallidum is a spirochaete bacterium with various subspecies that cause the diseases syphilis, bejel, and yaws. It is chiefly transmitted amongst humans, but it can also cause a condition found in cattle, bovine digital dermatitis. It is a helically coiled microorganism usually 6–15 μm long and 0.1–0.2 μm wide. T. pallidum's lack of either a tricarboxylic acid cycle or oxidative phosphorylation results in minimal metabolic activity. The treponemes have a cytoplasmic and an outer membrane. Using light microscopy, treponemes are visible only by using dark field illumination. Treponema pallidum consists of three subspecies, T. p. pallidum, T. p. endemicum, and T. p. pertenue, each of which has a distinct associated disease.

Neisseria is a large genus of bacteria that colonize the mucosal surfaces of many animals. Of the 11 species that colonize humans, only two are pathogens, N. meningitidis and N. gonorrhoeae. Most gonococcal infections are asymptomatic and self-resolving, and epidemic strains of the meningococcus may be carried in >95% of a population where systemic disease occurs at <1% prevalence.

Tularemia, also known as rabbit fever, is an infectious disease caused by the bacterium Francisella tularensis. Symptoms may include fever, skin ulcers, and enlarged lymph nodes. Occasionally, a form that results in pneumonia or a throat infection may occur.
In biology, immunity is the capability of multicellular organisms to resist harmful microorganisms. Immunity involves both specific and nonspecific components. The nonspecific components act as barriers or eliminators of a wide range of pathogens irrespective of their antigenic make-up. Other components of the immune system adapt themselves to each new disease encountered and can generate pathogen-specific immunity.

Chlamydia trachomatis, commonly known as chlamydia, is a bacterium that causes chlamydia, which can manifest in various ways, including: trachoma, lymphogranuloma venereum, nongonococcal urethritis, cervicitis, salpingitis, pelvic inflammatory disease. C. trachomatis is the most common infectious cause of blindness and the most common sexually transmitted bacterium.
Mycoplasma pneumoniae is a very small bacterium in the class Mollicutes. It is a human pathogen that causes the disease mycoplasma pneumonia, a form of atypical bacterial pneumonia related to cold agglutinin disease. M. pneumoniae is characterized by the absence of a peptidoglycan cell wall and resulting resistance to many antibacterial agents. The persistence of M. pneumoniae infections even after treatment is associated with its ability to mimic host cell surface composition.

Acinetobacter is a genus of gram-negative bacteria belonging to the wider class of Gammaproteobacteria. Acinetobacter species are oxidase-negative, exhibit twitching motility, and occur in pairs under magnification.

Haemophilus influenzae is a Gram-negative, coccobacillary, facultatively anaerobic capnophilic pathogenic bacterium of the family Pasteurellaceae. H. influenzae was first described in 1892 by Richard Pfeiffer during an influenza pandemic. He incorrectly described Haemophilus influenzae as the causative microbe, which retains "influenza" in its name.

Francisella is a genus of pathogenic, Gram-negative bacteria. They are small coccobacillary or rod-shaped, nonmotile organisms, which are also facultative intracellular parasites of macrophages. Strict aerobes, Francisella colonies bear a morphological resemblance to those of the genus Brucella.
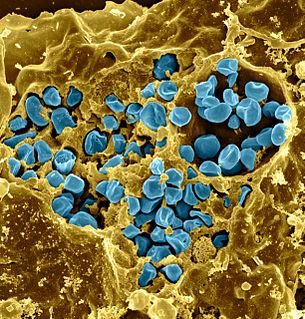
Francisella tularensis is a pathogenic species of Gram-negative coccobacillus, an aerobic bacterium. It is nonspore-forming, nonmotile, and the causative agent of tularemia, the pneumonic form of which is often lethal without treatment. It is a fastidious, facultative intracellular bacterium, which requires cysteine for growth. Due to its low infectious dose, ease of spread by aerosol, and high virulence, F. tularensis is classified as a Tier 1 Select Agent by the U.S. government, along with other potential agents of bioterrorism such as Yersinia pestis, Bacillus anthracis, and Ebola virus. When found in nature, Francisella tularensis can survive for several weeks at low temperatures in animal carcasses, soil, and water. In the laboratory, F. tularensis appears as small rods, and is grown best at 35–37 °C.
Rhinosporidium seeberi is a eukaryotic pathogen responsible for rhinosporidiosis, a disease which affects humans, horses, dogs, and to a lesser extent cattle, cats, foxes, and birds. It is most commonly found in tropical areas, especially India and Sri Lanka.
Influenza, commonly known as "the flu", is an infectious disease caused by influenza viruses. Symptoms range from mild to severe and often include fever, runny nose, sore throat, muscle pain, headache, coughing, and fatigue. These symptoms begin from one to four days after exposure to the virus and last for about 2–8 days. Diarrhea and vomiting can occur, particularly in children. Influenza may progress to pneumonia, which can be caused by the virus or by a subsequent bacterial infection. Other complications of infection include acute respiratory distress syndrome, meningitis, encephalitis, and worsening of pre-existing health problems such as asthma and cardiovascular disease.
Microbial toxins are toxins produced by micro-organisms, including bacteria, fungi, protozoa, dinoflagellates, and viruses. Many microbial toxins promote infection and disease by directly damaging host tissues and by disabling the immune system. Endotoxins most commonly refer to the lipopolysaccharide (LPS) or lipooligosaccharide (LOS) that are in the outer plasma membrane of Gram-negative bacteria. The botulinum toxin, which is primarily produced by Clostridium botulinum and less frequently by other Clostridium species, is the most toxic substance known in the world. However, microbial toxins also have important uses in medical science and research. Currently, new methods of detecting bacterial toxins are being developed to better isolate and understand these toxin. Potential applications of toxin research include combating microbial virulence, the development of novel anticancer drugs and other medicines, and the use of toxins as tools in neurobiology and cellular biology.
A neutralizing antibody (NAb) is an antibody that defends a cell from a pathogen or infectious particle by neutralizing any effect it has biologically. Neutralization renders the particle no longer infectious or pathogenic. Neutralizing antibodies are part of the humoral response of the adaptive immune system against viruses, intracellular bacteria and microbial toxin. By binding specifically to surface structures (antigen) on an infectious particle, neutralizing antibodies prevent the particle from interacting with its host cells it might infect and destroy. Immunity due to neutralizing antibodies is also known as sterilizing immunity, as the immune system eliminates the infectious particle before any infection takes place.

Escherichia coli is a gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms (endotherms). Most E. coli strains are harmless, but pathogenic varieties cause serious food poisoning, septic shock, meningitis, or urinary tract infections in humans. Unlike normal flora E. coli, the pathogenic varieties produce toxins and other virulence factors that enable them to reside in parts of the body normally not inhabited by E. coli, and to damage host cells. These pathogenic traits are encoded by virulence genes carried only by the pathogens.
This glossary of virology is a list of definitions of terms and concepts used in virology, the study of viruses, particularly in the description of viruses and their actions. Related fields include microbiology, molecular biology, and genetics.
Several small RNAs have been identified in Francisella tularensis, pathogenic bacterium that causes the disease tularaemia. Very little is known about Francisella's regulatory networks that allow this bacterium to survive in many environments.












