Related Research Articles
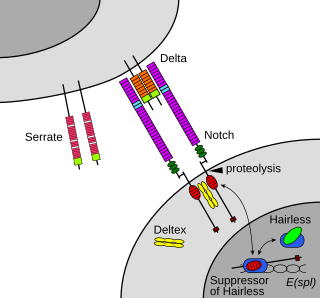
The Notch signaling pathway is a highly conserved cell signaling system present in most animals. Mammals possess four different notch receptors, referred to as NOTCH1, NOTCH2, NOTCH3, and NOTCH4. The notch receptor is a single-pass transmembrane receptor protein. It is a hetero-oligomer composed of a large extracellular portion, which associates in a calcium-dependent, non-covalent interaction with a smaller piece of the notch protein composed of a short extracellular region, a single transmembrane-pass, and a small intracellular region.

Neurogenic locus notch homolog protein 3(Notch 3) is a protein that in humans is encoded by the NOTCH3 gene.

Jagged1 (JAG1) is one of five cell surface proteins (ligands) that interact with four receptors in the mammalian Notch signaling pathway. The Notch Signaling Pathway is a highly conserved pathway that functions to establish and regulate cell fate decisions in many organ systems. Once the JAG1-NOTCH (receptor-ligand) interactions take place, a cascade of proteolytic cleavages is triggered resulting in activation of the transcription for downstream target genes. Located on human chromosome 20, the JAG1 gene is expressed in multiple organ systems in the body and causes the autosomal dominant disorder Alagille syndrome (ALGS) resulting from loss of function mutations within the gene. JAG1 has also been designated as CD339.

Neurogenic locus notch homolog protein 1(Notch 1) is a protein encoded in humans by the NOTCH1 gene. Notch 1 is a single-pass transmembrane receptor.

Neurogenic locus notch homolog 4(Notch 4) is a protein that in humans is encoded by the NOTCH4 gene located on chromosome 6.
Neurogenic locus notch homolog protein 2 is a protein that in humans is encoded by the NOTCH2 gene.

Delta-like protein 1 is a protein that in humans is encoded by the DLL1 gene.

Transcription factor HES1 is a protein that is encoded by the Hes1 gene, and is the mammalian homolog of the hairy gene in Drosophila. HES1 is one of the seven members of the Hes gene family (HES1-7). Hes genes code nuclear proteins that suppress transcription.

Hairy/enhancer-of-split related with YRPW motif protein 1 is a protein that in humans is encoded by the HEY1 gene.

Hairy/enhancer-of-split related with YRPW motif protein 2 (HEY2) also known as cardiovascular helix-loop-helix factor 1 (CHF1) is a protein that in humans is encoded by the HEY2 gene.

Beta-1,3-N-acetylglucosaminyltransferase manic fringe is an enzyme that in humans is encoded by the MFNG gene, a member of the fringe gene family which also includes the radical fringe (RFNG) and lunatic fringe (LFNG).

Protein atonal homolog 1 is a protein that in humans is encoded by the ATOH1 gene.

Protein deltex-1 is a protein that in humans is encoded by the DTX1 gene.

Beta-1,3-N-acetylglucosaminyltransferase lunatic fringe, (Lunatic Fringe), is a protein encoded in humans by the LFNG gene.

Transcription factor HES-5 is a protein that in humans is encoded by the HES5 gene.

Delta-like 3 (Drosophila), also known as DLL3, is a protein which in humans is encoded by the DLL3 gene. Two transcript variants encoding distinct isoforms have been identified for this gene.

Hairy/enhancer-of-split related with YRPW motif-like protein is a protein that in humans is encoded by the HEYL gene.

The STAT3-Ser/Hes3 signaling axis is a specific type of intracellular signaling pathway that regulates several fundamental properties of cells.

(HES7) or bHLHb37 is protein coding mammalian gene found on chromosome 17 in humans. HES7 is a member of the Hairy and Enhancer of Split families of Basic helix-loop-helix proteins. The gene product is a transcription factor and is expressed cyclically in the presomitic mesoderm as part of the Notch signalling pathway. HES7 is involved in the segmentation of somites from the presomitic mesoderm in vertebrates. The HES7 gene is self-regulated by a negative feedback loop in which the gene product can bind to its own promoter. This causes the gene to be expressed in an oscillatory manner. The HES7 protein also represses expression of Lunatic Fringe (LFNG) thereby both directly and indirectly regulating the Notch signalling pathway. Mutations in HES7 can result in deformities of the spine, ribs and heart. Spondylocostal dysostosis is a common disease caused by mutations in the HES7 gene. The inheritance pattern of Spondylocostal dysostosis is autosomal recessive.

Beta-1,3-N-acetylglucosaminyltransferase radical fringe, also known as radical fringe is a protein that in humans is encoded by the RFNG gene. Radical fringe is a signaling enzyme involved in the arrangement of the embryonic limb buds. It is a member of the fringe gene family, which also includes manic fringe and lunatic fringe. It is important for the dorsoventrally patterning of the limb and has been implicated in the formation of the apical ectodermal ridge (AER). The AER is essential for the distal patterning of the limb. Experiments executed in chicken models show Radical fringe is expressed in both the dorsal ectoderm and the AER. This provides evidence that the AER forms from cells already expressing radical fringe, though further evidence is needed to confirm. Grafting experiments have shown that formation of the AER comes from signals in the limb bud mesoderm. However, radical fringe acts as a permissive signal to create a boundary for the AER to form. The knockout experiments done in chicken models suggest Radical fringe plays an integral role in wing development.
References
- ↑ Gazave E, Lapébie P, Richards GS, Brunet F, Ereskovsky AV, Degnan BM, et al. (October 2009). "Origin and evolution of the Notch signalling pathway: an overview from eukaryotic genomes". BMC Evolutionary Biology. 9 (1): 249. Bibcode:2009BMCEE...9..249G. doi: 10.1186/1471-2148-9-249 . PMC 2770060 . PMID 19825158.
- ↑ Hopkin M (6 November 2006). "Troublesome gene names get the boot". Nature. doi: 10.1038/news061106-2 . S2CID 86514270.
- ↑ White M (26 September 2014). "Sonic Hedgehog, Dicer, and the Problem With Naming Genes". Pacific Standard. The Social Justice Foundation.