Related Research Articles

The polymerase chain reaction (PCR) is a method widely used to make millions to billions of copies of a specific DNA sample rapidly, allowing scientists to amplify a very small sample of DNA sufficiently to enable detailed study. PCR was invented in 1983 by American biochemist Kary Mullis at Cetus Corporation. Mullis and biochemist Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993.

A primer is a short, single-stranded nucleic acid used by all living organisms in the initiation of DNA synthesis. A synthetic primer may also be referred to as an oligo, short for oligonucleotide. DNA polymerase enzymes are only capable of adding nucleotides to the 3’-end of an existing nucleic acid, requiring a primer be bound to the template before DNA polymerase can begin a complementary strand. DNA polymerase adds nucleotides after binding to the RNA primer and synthesizes the whole strand. Later, the RNA strands must be removed accurately and replace them with DNA nucleotides forming a gap region known as a nick that is filled in using an enzyme called ligase. The removal process of the RNA primer requires several enzymes, such as Fen1, Lig1, and others that work in coordination with DNA polymerase, to ensure the removal of the RNA nucleotides and the addition of DNA nucleotides. Living organisms use solely RNA primers, while laboratory techniques in biochemistry and molecular biology that require in vitro DNA synthesis usually use DNA primers, since they are more temperature stable. Primers can be designed in laboratory for specific reactions such as polymerase chain reaction (PCR). When designing PCR primers, there are specific measures that must be taken into consideration, like the melting temperature of the primers and the annealing temperature of the reaction itself. Moreover, the DNA binding sequence of the primer in vitro has to be specifically chosen, which is done using a method called basic local alignment search tool (BLAST) that scans the DNA and finds specific and unique regions for the primer to bind.

DNA synthesis is the natural or artificial creation of deoxyribonucleic acid (DNA) molecules. DNA is a macromolecule made up of nucleotide units, which are linked by covalent bonds and hydrogen bonds, in a repeating structure. DNA synthesis occurs when these nucleotide units are joined to form DNA; this can occur artificially or naturally. Nucleotide units are made up of a nitrogenous base, pentose sugar (deoxyribose) and phosphate group. Each unit is joined when a covalent bond forms between its phosphate group and the pentose sugar of the next nucleotide, forming a sugar-phosphate backbone. DNA is a complementary, double stranded structure as specific base pairing occurs naturally when hydrogen bonds form between the nucleotide bases.
In molecular biology, an amplicon is a piece of DNA or RNA that is the source and/or product of amplification or replication events. It can be formed artificially, using various methods including polymerase chain reactions (PCR) or ligase chain reactions (LCR), or naturally through gene duplication. In this context, amplification refers to the production of one or more copies of a genetic fragment or target sequence, specifically the amplicon. As it refers to the product of an amplification reaction, amplicon is used interchangeably with common laboratory terms, such as "PCR product."
Xenobiology (XB) is a subfield of synthetic biology, the study of synthesizing and manipulating biological devices and systems. The name "xenobiology" derives from the Greek word xenos, which means "stranger, alien". Xenobiology is a form of biology that is not (yet) familiar to science and is not found in nature. In practice, it describes novel biological systems and biochemistries that differ from the canonical DNA–RNA-20 amino acid system. For example, instead of DNA or RNA, XB explores nucleic acid analogues, termed xeno nucleic acid (XNA) as information carriers. It also focuses on an expanded genetic code and the incorporation of non-proteinogenic amino acids, or “xeno amino acids” into proteins.
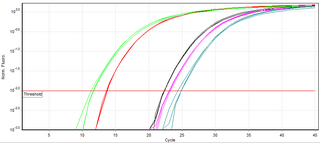
A real-time polymerase chain reaction is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR). It monitors the amplification of a targeted DNA molecule during the PCR, not at its end, as in conventional PCR. Real-time PCR can be used quantitatively and semi-quantitatively.
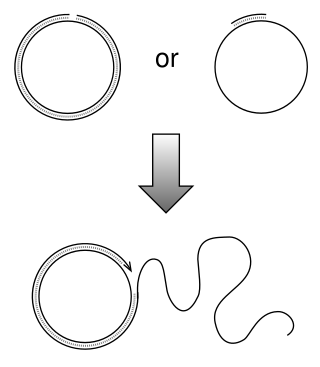
Rolling circle replication (RCR) is a process of unidirectional nucleic acid replication that can rapidly synthesize multiple copies of circular molecules of DNA or RNA, such as plasmids, the genomes of bacteriophages, and the circular RNA genome of viroids. Some eukaryotic viruses also replicate their DNA or RNA via the rolling circle mechanism.
GRE Subject Biochemistry, Cell and Molecular Biology was a standardized exam provided by ETS that was discontinued in December 2016. It is a paper-based exam and there are no computer-based versions of it. ETS places this exam three times per year: once in April, once in October and once in November. Some graduate programs in the United States recommend taking this exam, while others require this exam score as a part of the application to their graduate programs. ETS sends a bulletin with a sample practice test to each candidate after registration for the exam. There are 180 questions within the biochemistry subject test.
The following outline is provided as an overview of and topical guide to genetics:

A nucleic acid test (NAT) is a technique used to detect a particular nucleic acid sequence and thus usually to detect and identify a particular species or subspecies of organism, often a virus or bacterium that acts as a pathogen in blood, tissue, urine, etc. NATs differ from other tests in that they detect genetic materials rather than antigens or antibodies. Detection of genetic materials allows an early diagnosis of a disease because the detection of antigens and/or antibodies requires time for them to start appearing in the bloodstream. Since the amount of a certain genetic material is usually very small, many NATs include a step that amplifies the genetic material—that is, makes many copies of it. Such NATs are called nucleic acid amplification tests (NAATs). There are several ways of amplification, including polymerase chain reaction (PCR), strand displacement assay (SDA), transcription mediated assay (TMA), and loop-mediated isothermal amplification (LAMP).

The history of the polymerase chain reaction (PCR) has variously been described as a classic "Eureka!" moment, or as an example of cooperative teamwork between disparate researchers. Following is a list of events before, during, and after its development:
The versatility of polymerase chain reaction (PCR) has led to modifications of the basic protocol being used in a large number of variant techniques designed for various purposes. This article summarizes many of the most common variations currently or formerly used in molecular biology laboratories; familiarity with the fundamental premise by which PCR works and corresponding terms and concepts is necessary for understanding these variant techniques.
The ligase chain reaction (LCR) is a method of DNA amplification. The ligase chain reaction (LCR) is an amplification process that differs from polymerase chain reaction (PCR) in that it involves a thermostable ligase to join two probes or other molecules together which can then be amplified by standard PCR cycling. Each cycle results in a doubling of the target nucleic acid molecule. A key advantage of LCR is greater specificity as compared to PCR. Thus, LCR requires two completely different enzymes to operate properly: ligase, to join probe molecules together, and a thermostable polymerase (e.g., Taq polymerase) to amplify those molecules involved in successful ligation. The probes involved in the ligation are designed such that the 5′ end of one probe is directly adjacent to the 3′ end of the other probe, thereby providing the requisite 3′-OH and 5′-PO4 group substrates for the ligase.
Nicking Enzyme Amplification Reaction (NEAR) is a method for in vitro DNA amplification like the polymerase chain reaction (PCR). NEAR is isothermal, replicating DNA at a constant temperature using a polymerase to exponentially amplify the DNA at a temperature range of 55 °C to 59 °C.

Molecular cloning is a set of experimental methods in molecular biology that are used to assemble recombinant DNA molecules and to direct their replication within host organisms. The use of the word cloning refers to the fact that the method involves the replication of one molecule to produce a population of cells with identical DNA molecules. Molecular cloning generally uses DNA sequences from two different organisms: the species that is the source of the DNA to be cloned, and the species that will serve as the living host for replication of the recombinant DNA. Molecular cloning methods are central to many contemporary areas of modern biology and medicine.

Xenonucleic acids (XNAs) are synthetic nucleic acid analogues that are made up of non-natural components such as alternative nucleosides, sugars, or backbones.

Ligation is the joining of two nucleotides, or two nucleic acid fragments, into a single polymeric chain through the action of an enzyme known as a ligase. The reaction involves the formation of a phosphodiester bond between the 3'-hydroxyl terminus of one nucleotide and the 5'-phosphoryl terminus of another nucleotide, which results in the two nucleotides being linked consecutively on a single strand. Ligation works in fundamentally the same way for both DNA and RNA. A cofactor is generally involved in the reaction, usually ATP or NAD+. Eukaryotic ligases belong to the ATP type, while the NAD+ type are found in bacteria (e.g. E. coli).

Transcription-mediated amplification (TMA) is an isothermal, single-tube nucleic acid amplification system utilizing two enzymes, RNA polymerase and reverse transcriptase.
This glossary of cellular and molecular biology is a list of definitions of terms and concepts commonly used in the study of cell biology, molecular biology, and related disciplines, including molecular genetics, biochemistry, and microbiology. It is split across two articles:
This glossary of cellular and molecular biology is a list of definitions of terms and concepts commonly used in the study of cell biology, molecular biology, and related disciplines, including genetics, biochemistry, and microbiology. It is split across two articles:
References
- ↑ "Gene amplification - Latest research and news - Nature". www.nature.com.
- ↑ "PCR". Genetic Science Learning Center, University of Utah.
- ↑ Wiedmann, M (February 1994). "Ligase chain reaction (LCR) -- Overview and applications". PCR Methods and Applications. 3 (4): S51–64. doi: 10.1101/gr.3.4.s51 . PMID 8173509.
- ↑ Zhang J (2003). "Evolution by gene duplication: an update". Trends in Ecology & Evolution. 18 (6): 292–8. doi:10.1016/S0169-5347(03)00033-8.
- ↑ Graham Dellaire, Jason N Berman, Robert J. Arceci, eds., Cancer Genomics: From Bench to Personalized Medicine (2014), p. 205.