
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.

Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity.

Protein biosynthesis is a core biological process, occurring inside cells, balancing the loss of cellular proteins through the production of new proteins. Proteins perform a number of critical functions as enzymes, structural proteins or hormones. Protein synthesis is a very similar process for both prokaryotes and eukaryotes but there are some distinct differences.
A protein phosphatase is a phosphatase enzyme that removes a phosphate group from the phosphorylated amino acid residue of its substrate protein. Protein phosphorylation is one of the most common forms of reversible protein posttranslational modification (PTM), with up to 30% of all proteins being phosphorylated at any given time. Protein kinases (PKs) are the effectors of phosphorylation and catalyse the transfer of a γ-phosphate from ATP to specific amino acids on proteins. Several hundred PKs exist in mammals and are classified into distinct super-families. Proteins are phosphorylated predominantly on Ser, Thr and Tyr residues, which account for 79.3, 16.9 and 3.8% respectively of the phosphoproteome, at least in mammals. In contrast, protein phosphatases (PPs) are the primary effectors of dephosphorylation and can be grouped into three main classes based on sequence, structure and catalytic function. The largest class of PPs is the phosphoprotein phosphatase (PPP) family comprising PP1, PP2A, PP2B, PP4, PP5, PP6 and PP7, and the protein phosphatase Mg2+- or Mn2+-dependent (PPM) family, composed primarily of PP2C. The protein Tyr phosphatase (PTP) super-family forms the second group, and the aspartate-based protein phosphatases the third. The protein pseudophosphatases form part of the larger phosphatase family, and in most cases are thought to be catalytically inert, instead functioning as phosphate-binding proteins, integrators of signalling or subcellular traps. Examples of membrane-spanning protein phosphatases containing both active (phosphatase) and inactive (pseudophosphatase) domains linked in tandem are known, conceptually similar to the kinase and pseudokinase domain polypeptide structure of the JAK pseudokinases. A complete comparative analysis of human phosphatases and pseudophosphatases has been completed by Manning and colleagues, forming a companion piece to the ground-breaking analysis of the human kinome, which encodes the complete set of ~536 human protein kinases.

Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself or by forming a template for the production of proteins. RNA and deoxyribonucleic acid (DNA) are nucleic acids. The nucleic acids constitute one of the four major macromolecules essential for all known forms of life. RNA is assembled as a chain of nucleotides. Cellular organisms use messenger RNA (mRNA) to convey genetic information that directs synthesis of specific proteins. Many viruses encode their genetic information using an RNA genome.
The central dogma of molecular biology deals with the flow of genetic information within a biological system. It is often stated as "DNA makes RNA, and RNA makes protein", although this is not its original meaning. It was first stated by Francis Crick in 1957, then published in 1958:
The Central Dogma. This states that once "information" has passed into protein it cannot get out again. In more detail, the transfer of information from nucleic acid to nucleic acid, or from nucleic acid to protein may be possible, but transfer from protein to protein, or from protein to nucleic acid is impossible. Information here means the precise determination of sequence, either of bases in the nucleic acid or of amino acid residues in the protein.

A macromolecule is a very large molecule important to biological processes, such as a protein or nucleic acid. It is composed of thousands of covalently bonded atoms. Many macromolecules are polymers of smaller molecules called monomers. The most common macromolecules in biochemistry are biopolymers and large non-polymeric molecules such as lipids, nanogels and macrocycles. Synthetic fibers and experimental materials such as carbon nanotubes are also examples of macromolecules.

Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, proteins or non-coding RNA, and ultimately affect a phenotype. These products are often proteins, but in non-protein-coding genes such as transfer RNA (tRNA) and small nuclear RNA (snRNA), the product is a functional non-coding RNA. The process of gene expression is used by all known life—eukaryotes, prokaryotes, and utilized by viruses—to generate the macromolecular machinery for life.
Protein engineering is the process of developing useful or valuable proteins through the design and production of unnatural polypeptides, often by altering amino acid sequences found in nature. It is a young discipline, with much research taking place into the understanding of protein folding and recognition for protein design principles. It has been used to improve the function of many enzymes for industrial catalysis. It is also a product and services market, with an estimated value of $168 billion by 2017.

In biology, translation is the process in living cells in which proteins are produced using RNA molecules as templates. The generated protein is a sequence of amino acids. This sequence is determined by the sequence of nucleotides in the RNA. The nucleotides are considered three at a time. Each such triple results in addition of one specific amino acid to the protein being generated. The matching from nucleotide triple to amino acid is called the genetic code. The translation is performed by a large complex of functional RNA and proteins called ribosomes. The entire process is called gene expression.

A biomolecule or biological molecule is loosely defined as a molecule produced by a living organism and essential to one or more typically biological processes. Biomolecules include large macromolecules such as proteins, carbohydrates, lipids, and nucleic acids, as well as small molecules such as vitamins and hormones. A more general name for this class of material is biological materials. Biomolecules are an important element of living organisms, those biomolecules are often endogenous, produced within the organism but organisms usually need exogenous biomolecules, for example certain nutrients, to survive.

Silent mutations are mutations in DNA that do not have an observable effect on the organism's phenotype. They are a specific type of neutral mutation. The phrase silent mutation is often used interchangeably with the phrase synonymous mutation; however, synonymous mutations are not always silent, nor vice versa. Synonymous mutations can affect transcription, splicing, mRNA transport, and translation, any of which could alter phenotype, rendering the synonymous mutation non-silent. The substrate specificity of the tRNA to the rare codon can affect the timing of translation, and in turn the co-translational folding of the protein. This is reflected in the codon usage bias that is observed in many species. Mutations that cause the altered codon to produce an amino acid with similar functionality are often classified as silent; if the properties of the amino acid are conserved, this mutation does not usually significantly affect protein function.

In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and non-coding genes.

Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Rossmann fold for binding S-Adenosyl methionine (SAM). Class II methyltransferases contain a SET domain, which are exemplified by SET domain histone methyltransferases, and class III methyltransferases, which are membrane associated. Methyltransferases can also be grouped as different types utilizing different substrates in methyl transfer reactions. These types include protein methyltransferases, DNA/RNA methyltransferases, natural product methyltransferases, and non-SAM dependent methyltransferases. SAM is the classical methyl donor for methyltransferases, however, examples of other methyl donors are seen in nature. The general mechanism for methyl transfer is a SN2-like nucleophilic attack where the methionine sulfur serves as the leaving group and the methyl group attached to it acts as the electrophile that transfers the methyl group to the enzyme substrate. SAM is converted to S-Adenosyl homocysteine (SAH) during this process. The breaking of the SAM-methyl bond and the formation of the substrate-methyl bond happen nearly simultaneously. These enzymatic reactions are found in many pathways and are implicated in genetic diseases, cancer, and metabolic diseases. Another type of methyl transfer is the radical S-Adenosyl methionine (SAM) which is the methylation of unactivated carbon atoms in primary metabolites, proteins, lipids, and RNA.

Artificial transcription factors (ATFs) are engineered individual or multi molecule transcription factors that either activate or repress gene transcription (biology).
Protein metabolism denotes the various biochemical processes responsible for the synthesis of proteins and amino acids (anabolism), and the breakdown of proteins by catabolism.
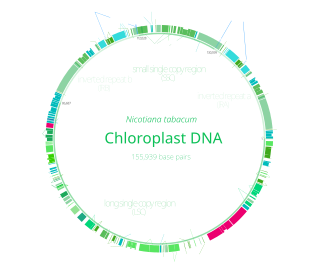
Chloroplast DNA (cpDNA) is the DNA located in chloroplasts, which are photosynthetic organelles located within the cells of some eukaryotic organisms. Chloroplasts, like other types of plastid, contain a genome separate from that in the cell nucleus. The existence of chloroplast DNA was identified biochemically in 1959, and confirmed by electron microscopy in 1962. The discoveries that the chloroplast contains ribosomes and performs protein synthesis revealed that the chloroplast is genetically semi-autonomous. The first complete chloroplast genome sequences were published in 1986, Nicotiana tabacum (tobacco) by Sugiura and colleagues and Marchantia polymorpha (liverwort) by Ozeki et al. Since then, a great number of chloroplast DNAs from various species have been sequenced.
Numerous key discoveries in biology have emerged from studies of RNA, including seminal work in the fields of biochemistry, genetics, microbiology, molecular biology, molecular evolution, and structural biology. As of 2010, 30 scientists have been awarded Nobel Prizes for experimental work that includes studies of RNA. Specific discoveries of high biological significance are discussed in this article.
This glossary of cellular and molecular biology is a list of definitions of terms and concepts commonly used in the study of cell biology, molecular biology, and related disciplines, including molecular genetics, biochemistry, and microbiology. It is split across two articles:
This glossary of cellular and molecular biology is a list of definitions of terms and concepts commonly used in the study of cell biology, molecular biology, and related disciplines, including genetics, biochemistry, and microbiology. It is split across two articles:









