Chromatin is a complex of DNA and protein found in eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important roles in reinforcing the DNA during cell division, preventing DNA damage, and regulating gene expression and DNA replication. During mitosis and meiosis, chromatin facilitates proper segregation of the chromosomes in anaphase; the characteristic shapes of chromosomes visible during this stage are the result of DNA being coiled into highly condensed chromatin.

In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei and in most Archaeal phyla. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn are wrapped into 30-nanometer fibers that form tightly packed chromatin. Histones prevent DNA from becoming tangled and protect it from DNA damage. In addition, histones play important roles in gene regulation and DNA replication. Without histones, unwound DNA in chromosomes would be very long. For example, each human cell has about 1.8 meters of DNA if completely stretched out; however, when wound about histones, this length is reduced to about 90 micrometers (0.09 mm) of 30 nm diameter chromatin fibers.

In biology, epigenetics is the study of heritable traits, or a stable change of cell function, that happen without changes to the DNA sequence. The Greek prefix epi- in epigenetics implies features that are "on top of" or "in addition to" the traditional genetic mechanism of inheritance. Epigenetics usually involves a change that is not erased by cell division, and affects the regulation of gene expression. Such effects on cellular and physiological phenotypic traits may result from environmental factors, or be part of normal development. They can lead to cancer.
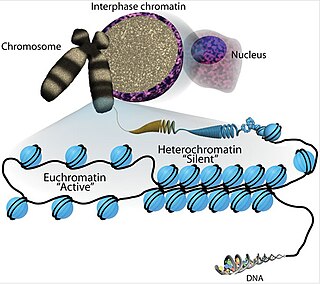
Euchromatin is a lightly packed form of chromatin that is enriched in genes, and is often under active transcription. Euchromatin stands in contrast to heterochromatin, which is tightly packed and less accessible for transcription. 92% of the human genome is euchromatic.

Histone methyltransferases (HMT) are histone-modifying enzymes, that catalyze the transfer of one, two, or three methyl groups to lysine and arginine residues of histone proteins. The attachment of methyl groups occurs predominantly at specific lysine or arginine residues on histones H3 and H4. Two major types of histone methyltranferases exist, lysine-specific and arginine-specific. In both types of histone methyltransferases, S-Adenosyl methionine (SAM) serves as a cofactor and methyl donor group.
The genomic DNA of eukaryotes associates with histones to form chromatin. The level of chromatin compaction depends heavily on histone methylation and other post-translational modifications of histones. Histone methylation is a principal epigenetic modification of chromatin that determines gene expression, genomic stability, stem cell maturation, cell lineage development, genetic imprinting, DNA methylation, and cell mitosis.

Histone H3 is one of the five main histones involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal tail, H3 is involved with the structure of the nucleosomes of the 'beads on a string' structure. Histone proteins are highly post-translationally modified however Histone H3 is the most extensively modified of the five histones. The term "Histone H3" alone is purposely ambiguous in that it does not distinguish between sequence variants or modification state. Histone H3 is an important protein in the emerging field of epigenetics, where its sequence variants and variable modification states are thought to play a role in the dynamic and long term regulation of genes.

Histone H2A is one of the five main histone proteins involved in the structure of chromatin in eukaryotic cells.
Histone H2B is one of the 5 main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and long N-terminal and C-terminal tails, H2B is involved with the structure of the nucleosomes.

Citrullination or deimination is the conversion of the amino acid arginine in a protein into the amino acid citrulline. Citrulline is not one of the 20 standard amino acids encoded by DNA in the genetic code. Instead, it is the result of a post-translational modification. Citrullination is distinct from the formation of the free amino acid citrulline as part of the urea cycle or as a byproduct of enzymes of the nitric oxide synthase family.

Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Rossmann fold for binding S-Adenosyl methionine (SAM). Class II methyltransferases contain a SET domain, which are exemplified by SET domain histone methyltransferases, and class III methyltransferases, which are membrane associated. Methyltransferases can also be grouped as different types utilizing different substrates in methyl transfer reactions. These types include protein methyltransferases, DNA/RNA methyltransferases, natural product methyltransferases, and non-SAM dependent methyltransferases. SAM is the classical methyl donor for methyltransferases, however, examples of other methyl donors are seen in nature. The general mechanism for methyl transfer is a SN2-like nucleophilic attack where the methionine sulfur serves as the leaving group and the methyl group attached to it acts as the electrophile that transfers the methyl group to the enzyme substrate. SAM is converted to S-Adenosyl homocysteine (SAH) during this process. The breaking of the SAM-methyl bond and the formation of the substrate-methyl bond happen nearly simultaneously. These enzymatic reactions are found in many pathways and are implicated in genetic diseases, cancer, and metabolic diseases. Another type of methyl transfer is the radical S-Adenosyl methionine (SAM) which is the methylation of unactivated carbon atoms in primary metabolites, proteins, lipids, and RNA.
The histone code is a hypothesis that the transcription of genetic information encoded in DNA is in part regulated by chemical modifications to histone proteins, primarily on their unstructured ends. Together with similar modifications such as DNA methylation it is part of the epigenetic code. Histones associate with DNA to form nucleosomes, which themselves bundle to form chromatin fibers, which in turn make up the more familiar chromosome. Histones are globular proteins with a flexible N-terminus that protrudes from the nucleosome. Many of the histone tail modifications correlate very well to chromatin structure and both histone modification state and chromatin structure correlate well to gene expression levels. The critical concept of the histone code hypothesis is that the histone modifications serve to recruit other proteins by specific recognition of the modified histone via protein domains specialized for such purposes, rather than through simply stabilizing or destabilizing the interaction between histone and the underlying DNA. These recruited proteins then act to alter chromatin structure actively or to promote transcription. For details of gene expression regulation by histone modifications see table below.

Acetyltransferase is a type of transferase enzyme that transfers an acetyl group, through a process called acetylation. Acetylation serves as a modification that can profoundly transform the functionality of a protein by modifying various properties like hydrophobicity, solubility, and surface attributes. These alterations have the potential to influence the protein's conformation and its interactions with substrates, cofactors, and other macromolecules. The image to the right shows the basic structure of an acetyl group, where R is a variable indicates the remainder of the molecule to which the acetyl group is attached.

Histone-modifying enzymes are enzymes involved in the modification of histone substrates after protein translation and affect cellular processes including gene expression. To safely store the eukaryotic genome, DNA is wrapped around four core histone proteins, which then join to form nucleosomes. These nucleosomes further fold together into highly condensed chromatin, which renders the organism's genetic material far less accessible to the factors required for gene transcription, DNA replication, recombination and repair. Subsequently, eukaryotic organisms have developed intricate mechanisms to overcome this repressive barrier imposed by the chromatin through histone modification, a type of post-translational modification which typically involves covalently attaching certain groups to histone residues. Once added to the histone, these groups elicit either a loose and open histone conformation, euchromatin, or a tight and closed histone conformation, heterochromatin. Euchromatin marks active transcription and gene expression, as the light packing of histones in this way allows entry for proteins involved in the transcription process. As such, the tightly packed heterochromatin marks the absence of current gene expression.
Chromatin remodeling is the dynamic modification of chromatin architecture to allow access of condensed genomic DNA to the regulatory transcription machinery proteins, and thereby control gene expression. Such remodeling is principally carried out by 1) covalent histone modifications by specific enzymes, e.g., histone acetyltransferases (HATs), deacetylases, methyltransferases, and kinases, and 2) ATP-dependent chromatin remodeling complexes which either move, eject or restructure nucleosomes. Besides actively regulating gene expression, dynamic remodeling of chromatin imparts an epigenetic regulatory role in several key biological processes, egg cells DNA replication and repair; apoptosis; chromosome segregation as well as development and pluripotency. Aberrations in chromatin remodeling proteins are found to be associated with human diseases, including cancer. Targeting chromatin remodeling pathways is currently evolving as a major therapeutic strategy in the treatment of several cancers.

Euchromatic histone-lysine N-methyltransferase 2 (EHMT2), also known as G9a, is a histone methyltransferase enzyme that in humans is encoded by the EHMT2 gene. G9a deposits the mono- and di-methylated states of histone H3 at lysine residue 9 and lysine residue 27. The presence of H3K9me1/2 is usually associated with gene silencing.
Trithorax-group proteins (TrxG) are a heterogeneous collection of proteins whose main action is to maintain gene expression. They can be categorized into three general classes based on molecular function:
- histone-modifying TrxG proteins
- chromatin-remodeling TrxG proteins
- DNA-binding TrxG proteins,
H3K27ac is an epigenetic modification to the DNA packaging protein histone H3. It is a mark that indicates acetylation of the lysine residue at N-terminal position 27 of the histone H3 protein.
H4K5ac is an epigenetic modification to the DNA packaging protein histone H4. It is a mark that indicates the acetylation at the 5th lysine residue of the histone H4 protein. H4K5 is the closest lysine residue to the N-terminal tail of histone H4. It is enriched at the transcription start site (TSS) and along gene bodies. Acetylation of histone H4K5 and H4K12ac is enriched at centromeres.
H4K8ac, representing an epigenetic modification to the DNA packaging protein histone H4, is a mark indicating the acetylation at the 8th lysine residue of the histone H4 protein. It has been implicated in the prevalence of malaria.
H3T3P is an epigenetic modification to the DNA packaging protein histone H3. It is a mark that indicates the phosphorylation the 3rd threonine residue of the histone H3 protein.










