
In computing, Common Gateway Interface (CGI) is an interface specification that enables web servers to execute an external program to process HTTP or HTTPS user requests.
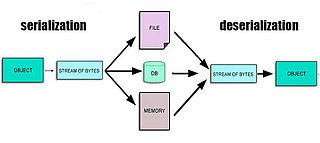
In computing, serialization is the process of translating a data structure or object state into a format that can be stored or transmitted and reconstructed later. When the resulting series of bits is reread according to the serialization format, it can be used to create a semantically identical clone of the original object. For many complex objects, such as those that make extensive use of references, this process is not straightforward. Serialization of objects does not include any of their associated methods with which they were previously linked.
GNUstep is a free software implementation of the Cocoa Objective-C frameworks, widget toolkit, and application development tools for Unix-like operating systems and Microsoft Windows. It is part of the GNU Project.

The Biopython project is an open-source collection of non-commercial Python tools for computational biology and bioinformatics, created by an international association of developers. It contains classes to represent biological sequences and sequence annotations, and it is able to read and write to a variety of file formats. It also allows for a programmatic means of accessing online databases of biological information, such as those at NCBI. Separate modules extend Biopython's capabilities to sequence alignment, protein structure, population genetics, phylogenetics, sequence motifs, and machine learning. Biopython is one of a number of Bio* projects designed to reduce code duplication in computational biology.

PyQt is a Python binding of the cross-platform GUI toolkit Qt, implemented as a Python plug-in. PyQt is free software developed by the British firm Riverbank Computing. It is available under similar terms to Qt versions older than 4.5; this means a variety of licenses including GNU General Public License (GPL) and commercial license, but not the GNU Lesser General Public License (LGPL). PyQt supports Microsoft Windows as well as various kinds of UNIX, including Linux and MacOS.
Perl Data Language is a set of free software array programming extensions to the Perl programming language. PDL extends the data structures built into Perl, to include large multidimensional arrays, and adds functionality to manipulate those arrays as vector objects. It also provides tools for image processing, machine learning, computer modeling of physical systems, and graphical plotting and presentation. Simple operations are automatically vectorized across complete arrays, and higher-dimensional operations are supported.

ROOT is an object-oriented computer program and library developed by CERN. It was originally designed for particle physics data analysis and contains several features specific to the field, but it is also used in other applications such as astronomy and data mining. The latest minor release is 6.32, as of 2024-05-26.
Paul Kunz was an American particle physicist and software developer, who initiated the deployment of the first web server outside of Europe. After a meeting in September 1991 with Tim Berners-Lee of CERN, he returned to the Stanford Linear Accelerator Center (SLAC) with word of the World Wide Web. By Thursday, December 12, 1991, there was an active Web server installed and operational at SLAC, establishing the first Web server in the US, the SPIRES HEP, connected to the SPIRES High Energy Physics database, thanks to the efforts of Kunz, Louise Addis, and Terry Hung.
In the field of computer network administration, pcap is an application programming interface (API) for capturing network traffic. While the name is an abbreviation of packet capture, that is not the API's proper name. Unix-like systems implement pcap in the libpcap library; for Windows, there is a port of libpcap named WinPcap that is no longer supported or developed, and a port named Npcap for Windows 7 and later that is still supported.
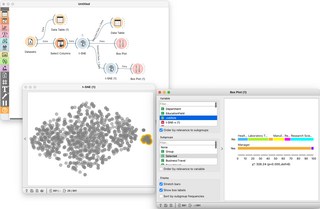
Orange is an open-source data visualization, machine learning and data mining toolkit. It features a visual programming front-end for exploratory qualitative data analysis and interactive data visualization.
CellProfiler is free, open-source software designed to enable biologists without training in computer vision or programming to quantitatively measure phenotypes from thousands of images automatically. Advanced algorithms for image analysis are available as individual modules that can be placed in sequential order together to form a pipeline; the pipeline is then used to identify and measure biological objects and features in images, particularly those obtained through fluorescence microscopy.
Roundup is an open-source issue or bug tracking system featuring a command-line, web and e-mail interface. It is written in Python and designed to be highly customizable.
NetCDF is a set of software libraries and self-describing, machine-independent data formats that support the creation, access, and sharing of array-oriented scientific data. The project homepage is hosted by the Unidata program at the University Corporation for Atmospheric Research (UCAR). They are also the chief source of netCDF software, standards development, updates, etc. The format is an open standard. NetCDF Classic and 64-bit Offset Format are an international standard of the Open Geospatial Consortium.

Shinken is an open source computer system and network monitoring software application compatible with Nagios. It watches hosts and services, gathers performance data and alerts users when error conditions occur and again when the conditions clear.
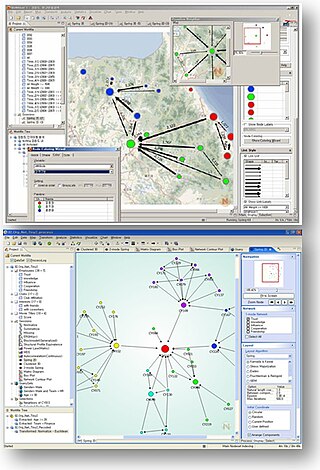
NetMiner is an application software for exploratory analysis and visualization of large network data based on SNA. It can be used for general research and teaching in social networks. This tool allows researchers to explore their network data visually and interactively, helps them to detect underlying patterns and structures of the network. It features data transformation, network analysis, statistics, visualization of network data, chart, and a programming language based on the Python script language. Also, it enables users to import unstructured text data(e.g. news, articles, tweets, etc.) and extract words and network from text data. In addition, NetMiner SNS Data Collector, an extension of NetMiner, can collect some social networking service(SNS) data with a few clicks.

Nim is a general-purpose, multi-paradigm, statically typed, compiled high-level system programming language, designed and developed by a team around Andreas Rumpf. Nim is designed to be "efficient, expressive, and elegant", supporting metaprogramming, functional, message passing, procedural, and object-oriented programming styles by providing several features such as compile time code generation, algebraic data types, a foreign function interface (FFI) with C, C++, Objective-C, and JavaScript, and supporting compiling to those same languages as intermediate representations.
BisQue is a free, open source web-based platform for the exchange and exploration of large, complex datasets. It is being developed at the Vision Research Lab at the University of California, Santa Barbara. BisQue specifically supports large scale, multi-dimensional multimodal-images and image analysis. Metadata is stored as arbitrarily nested and linked tag/value pairs, allowing for domain-specific data organization. Image analysis modules can be added to perform complex analysis tasks on compute clusters. Analysis results are stored within the database for further querying and processing. The data and analysis provenance is maintained for reproducibility of results. BisQue can be easily deployed in cloud computing environments or on computer clusters for scalability. BisQue has been integrated into the NSF Cyberinfrastructure project CyVerse. The user interacts with BisQue via any modern web browser.
MicroPython is a software implementation of a programming language largely compatible with Python 3, written in C, that is optimized to run on a microcontroller.










