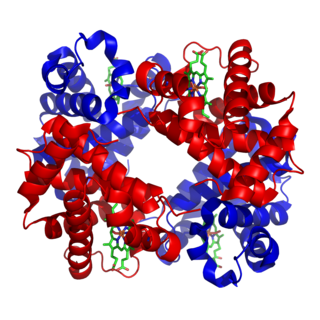
Metalloprotein is a generic term for a protein that contains a metal ion cofactor. A large proportion of all proteins are part of this category. For instance, at least 1000 human proteins contain zinc-binding protein domains although there may be up to 3000 human zinc metalloproteins.

Exodeoxyribonuclease V is an enzyme of E. coli that initiates recombinational repair from potentially lethal double strand breaks in DNA which may result from ionizing radiation, replication errors, endonucleases, oxidative damage, and a host of other factors. The RecBCD enzyme is both a helicase that unwinds, or separates the strands of DNA, and a nuclease that makes single-stranded nicks in DNA. It catalyses exonucleolytic cleavage in either 5′- to 3′- or 3′- to 5′-direction to yield 5′-phosphooligonucleotides.
A hydrogenase is an enzyme that catalyses the reversible oxidation of molecular hydrogen (H2), as shown below:

Endopeptidase Clp (EC 3.4.21.92, endopeptidase Ti, caseinolytic protease, protease Ti, ATP-dependent Clp protease, ClpP, Clp protease). This enzyme catalyses the following chemical reaction
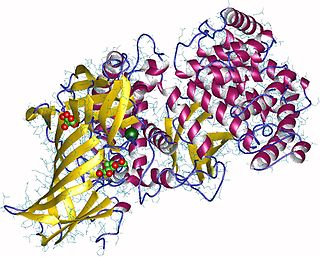
Aminopeptidases are enzymes that catalyze the cleavage of amino acids from the amino terminus (N-terminus) of proteins or peptides (exopeptidases). They are widely distributed throughout the animal and plant kingdoms and are found in many subcellular organelles, in cytosol, and as membrane components. Aminopeptidases are used in essential cellular functions. Many, but not all, of these peptidases are zinc metalloenzymes.
Bioorganometallic chemistry is the study of biologically active molecules that contain carbon directly bonded to metals or metalloids. The importance of main-group and transition-metal centers has long been recognized as important to the function of enzymes and other biomolecules. However, only a small subset of naturally-occurring metal complexes and synthetically prepared pharmaceuticals are organometallic; that is, they feature a direct covalent bond between the metal(loid) and a carbon atom. The first, and for a long time, the only examples of naturally occurring bioorganometallic compounds were the cobalamin cofactors (vitamin B12) in its various forms. In the 21st century, discovery of new systems containing carbon-metal bonds in biology, bioorganometallic chemistry is rapidly emerging as a distinct subdiscipline of bioinorganic chemistry that straddles organometallic chemistry and biochemistry. Naturally occurring bioorganometallics include enzymes and sensor proteins. Also within this realm are synthetically prepared organometallic compounds that serve as new drugs and imaging agents (technetium-99m sestamibi) as well as the principles relevant to the toxicology of organometallic compounds (e.g., methylmercury). Consequently, bioorganometallic chemistry is increasingly relevant to medicine and pharmacology.
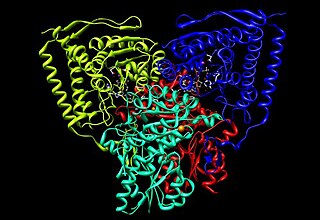
Pyruvate dehydrogenase is an enzyme that catalyzes the reaction of pyruvate and a lipoamide to give the acetylated dihydrolipoamide and carbon dioxide. The conversion requires the coenzyme thiamine pyrophosphate.

Cupriavidus necator is a Gram-negative soil bacterium of the class Betaproteobacteria.
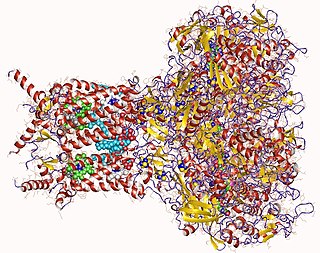
Formate dehydrogenases are a set of enzymes that catalyse the oxidation of formate to carbon dioxide, donating the electrons to a second substrate, such as NAD+ in formate:NAD+ oxidoreductase (EC 1.17.1.9) or to a cytochrome in formate:ferricytochrome-b1 oxidoreductase (EC 1.2.2.1). This family of enzymes has attracted attention as inspiration or guidance on methods for the carbon dioxide fixation, relevant to global warming.
In enzymology, carbon monoxide dehydrogenase (CODH) (EC 1.2.7.4) is an enzyme that catalyzes the chemical reaction
In enzymology, a hydrogen:quinone oxidoreductase (EC 1.12.5.1) is an enzyme that catalyzes the chemical reaction

ATP-dependent Clp protease proteolytic subunit (ClpP) is an enzyme that in humans is encoded by the CLPP gene. This protein is an essential component to form the protein complex of Clp protease.
Hydrogenases are enzymes that catalyze the reversible activation of hydrogen and which occur widely in prokaryotes as well as in some eukaryotes. There are various types of hydrogenases, but all of them seem to contain at least one iron-sulphur cluster. They can be broadly divided into two groups: hydrogenases containing nickel and, in some cases, also selenium and those lacking nickel.
[NiFe] hydrogenase is a type of hydrogenase, which is an oxidative enzyme that reversibly converts molecular hydrogen in prokaryotes including Bacteria and Archaea. The catalytic site on the enzyme provides simple hydrogen-metabolizing microorganisms a redox mechanism by which to store and utilize energy via the reaction
Ribonuclease E is a bacterial ribonuclease that participates in the processing of ribosomal RNA and the chemical degradation of bulk cellular RNA.
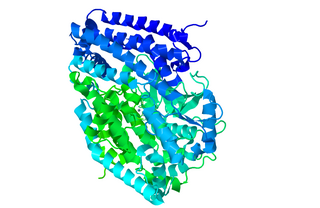
Peptidyl-dipeptidase Dcp (EC 3.4.15.5, dipeptidyl carboxypeptidase (Dcp), dipeptidyl carboxypeptidase) is a metalloenzyme found in the cytoplasm of bacterium E. Coli responsible for the C-terminal cleavage of a variety of dipeptides and unprotected larger peptide chains. The enzyme does not hydrolyze bonds in which P1' is Proline, or both P1 and P1' are Glycine. Dcp consists of 680 amino acid residues that form into a single active monomer which aids in the intracellular degradation of peptides. Dcp coordinates to divalent zinc which sits in the pocket of the active site and is composed of four subsites: S1’, S1, S2, and S3, each subsite attracts certain amino acids at a specific position on the substrate enhancing the selectivity of the enzyme. The four subsites detect and bind different amino acid types on the substrate peptide in the P1 and P2 positions. Some metallic divalent cations such as Ni+2, Cu+2, and Zn+2 inhibit the function of the enzyme around 90%, whereas other cations such as Mn+2, Ca+2, Mg+2, and Co+2 have slight catalyzing properties, and increase the function by around 20%. Basic amino acids such as Arginine bind preferably at the S1 site, the S2 site sits deeper in the enzyme therefore is restricted to bind hydrophobic amino acids with phenylalanine in the P2 position. Dcp is divided into two subdomains (I, and II), which are the two sides of the clam shell-like structure and has a deep inner cavity where a pair of histidine residues bind to the catalytic zinc ion in the active site. Peptidyl-Dipeptidase Dcp is classified like Angiotensin-I converting enzyme (ACE) which is also a carboxypeptidase involved in blood pressure regulation, but due to structural differences and peptidase activity between these two enzymes they had to be examined separately. ACE has endopeptidase activity, whereas Dcp strictly has exopeptidase activity based on its cytoplasmic location and therefore their mechanisms of action are differentiated. Another difference between these enzymes is that the activity of Peptidyl-Dipeptidase Dcp is not enhanced in the presence of chloride anions, whereas chloride enhances ACE activity.

Peptidase Do is an enzyme. This enzyme catalyses the following chemical reaction
HycI peptidase is an enzyme. This enzyme catalyses the following chemical reaction

Acetyl-CoA synthase (ACS), not to be confused with Acetyl-CoA synthetase or Acetate-CoA ligase, is a nickel-containing enzyme involved in the metabolic processes of cells. Together with Carbon monoxide dehydrogenase (CODH), it forms the bifunctional enzyme Acetyl-CoA Synthase/Carbon Monoxide Dehydrogenase (ACS/CODH) found in anaerobic organisms such as archaea and bacteria. The ACS/CODH enzyme works primarily through the Wood–Ljungdahl pathway which converts carbon dioxide to Acetyl-CoA. The recommended name for this enzyme is CO-methylating acetyl-CoA synthase.
Proteins currently known to belong to the Ni2+-Co2+ Transporter (NiCoT) family (TC# 2.A.52) can be found in organisms ranging from Gram-negative and Gram-positive bacteria to archaea and some eukaryotes. Members of this family catalyze uptake of Ni2+ and/or Co2+ in a proton motive force-dependent process.











