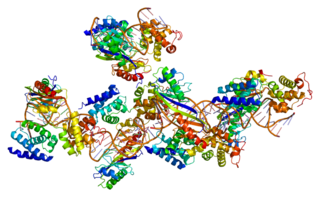
Ribosomes ( ) are macromolecular machines, found within all cells, that perform biological protein synthesis. Ribosomes link amino acids together in the order specified by the codons of messenger RNA (mRNA) molecules to form polypeptide chains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or more ribosomal RNA (rRNA) molecules and many ribosomal proteins. The ribosomes and associated molecules are also known as the translational apparatus.

Microbial ecology is the ecology of microorganisms: their relationship with one another and with their environment. It concerns the three major domains of life—Eukaryota, Archaea, and Bacteria—as well as viruses.
The Shine–Dalgarno (SD) sequence is a ribosomal binding site in bacterial and archaeal messenger RNA, generally located around 8 bases upstream of the start codon AUG. The RNA sequence helps recruit the ribosome to the messenger RNA (mRNA) to initiate protein synthesis by aligning the ribosome with the start codon. Once recruited, tRNA may add amino acids in sequence as dictated by the codons, moving downstream from the translational start site.

Metagenomics is the study of genetic material recovered directly from environmental or clinical samples by a method called sequencing. The broad field may also be referred to as environmental genomics, ecogenomics, community genomics or microbiomics.

Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosomal DNA (rDNA) and then bound to ribosomal proteins to form small and large ribosome subunits. rRNA is the physical and mechanical factor of the ribosome that forces transfer RNA (tRNA) and messenger RNA (mRNA) to process and translate the latter into proteins. Ribosomal RNA is the predominant form of RNA found in most cells; it makes up about 80% of cellular RNA despite never being translated into proteins itself. Ribosomes are composed of approximately 60% rRNA and 40% ribosomal proteins by mass.
Bacterial translation is the process by which messenger RNA is translated into proteins in bacteria.

The trp operon is a group of genes that are transcribed together, encoding the enzymes that produce the amino acid tryptophan in bacteria. The trp operon was first characterized in Escherichia coli, and it has since been discovered in many other bacteria. The operon is regulated so that, when tryptophan is present in the environment, the genes for tryptophan synthesis are repressed.

George Edward Fox is an astrobiologist, a Professor Emeritus and researcher at the University of Houston. He is an elected fellow of the American Academy of Microbiology, the American Association for the Advancement of Science, American Institute for Medical and Biological Engineering and the International Astrobiology Society. Fox received his B.A. degree in 1967, and completed his Ph.D. degree in 1974; both in chemical engineering at Syracuse University.
Transcription factor II B (TFIIB) is a general transcription factor that is involved in the formation of the RNA polymerase II preinitiation complex (PIC) and aids in stimulating transcription initiation. TFIIB is localised to the nucleus and provides a platform for PIC formation by binding and stabilising the DNA-TBP complex and by recruiting RNA polymerase II and other transcription factors. It is encoded by the TFIIB gene, and is homologous to archaeal transcription factor B and analogous to bacterial sigma factors.

16S ribosomal RNA is the RNA component of the 30S subunit of a prokaryotic ribosome. It binds to the Shine-Dalgarno sequence and provides most of the SSU structure.

Archaea is a domain of single-celled organisms. These microorganisms lack cell nuclei and are therefore prokaryotes. Archaea were initially classified as bacteria, receiving the name archaebacteria, but this term has fallen out of use.

Microbiota are the range of microorganisms that may be commensal, symbiotic, or pathogenic found in and on all multicellular organisms, including plants. Microbiota include bacteria, archaea, protists, fungi, and viruses, and have been found to be crucial for immunologic, hormonal, and metabolic homeostasis of their host.
HNH Endonuclease-Associated RNA and ORF (HEARO) RNAs conform to a conserved RNA structure that was identified in bacteria by bioinformatics. HEARO RNAs average roughly 300 nucleotides, which is comparable to the size of many ribozymes, which catalyze chemical reactions.

A wide variety of non-coding RNAs have been identified in various species of organisms known to science. However, RNAs have also been identified in "metagenomics" sequences derived from samples of DNA or RNA extracted from the environment, which contain unknown species. Initial work in this area detected homologs of known bacterial RNAs in such metagenome samples. Many of these RNA sequences were distinct from sequences within cultivated bacteria, and provide the potential for additional information on the RNA classes to which they belong.
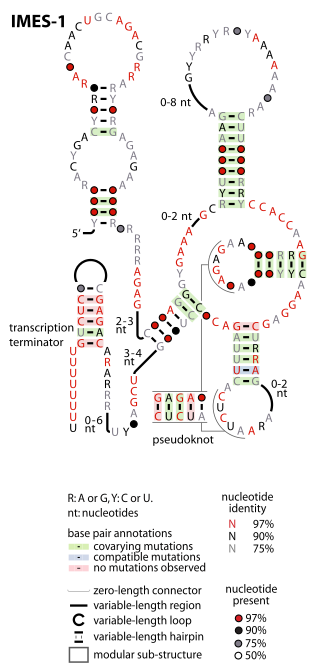
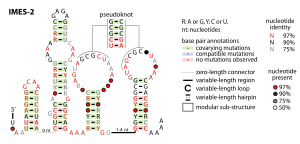
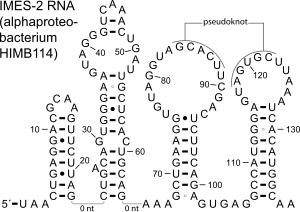
The IMES-1 RNA motif is a conserved RNA structure that was identified in marine environmental sequences by two studies based on metagenomics and bioinformatics, the first analyzing metatranscriptome (RNA) data and the second using metagenome (DNA) data. These RNAs are present in environmental sequences, and as of 2009 are not known to be present in any cultivated species. However, the species that use these RNAs are most closely related to known alphaproteobacteria and gammaproteobacteria. IMES-1 RNAs make up a significant portion of marine RNA transcripts and are exceptionally abundant in that over five times as many IMES-1 RNAs were found as ribosomes in RNAs sampled from the Pacific Ocean. Only two bacterial RNAs are known to be more highly transcribed than ribosomes. IMES-1 RNAs were also detected in abundance in Block Island Sound in the Atlantic Ocean.

The IMES-3 RNA motif is a conserved RNA structure that was identified based on metagenomics and bioinformatics, and the underlying RNA sequences were identified independently by an earlier study. These RNAs are present in environmental sequences, and as of 2009 are not known to be present in any cultivated species. IMES-3 RNAs are abundant in comparison to ribosomes in RNAs sampled from the Pacific Ocean.
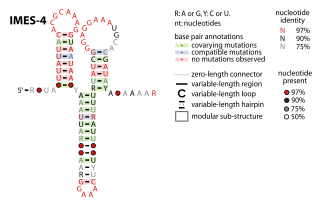
The IMES-4 RNA motif is a conserved RNA structure that was identified in marine environmental sequences by metagenomics and bioinformatics. These RNAs are present in environmental sequences, and as of 2009 are not known to be present in any cultivated species. IMES-4 RNAs are fairly abundant in comparison to ribosomes in RNAs sampled from the Pacific Ocean.

The Gut-1 RNA motif is a conserved RNA structure identified by bioinformatics. These RNAs are present in environmental sequences, and as of 2010 are not known to be present in any species that has been grown under laboratory conditions. Gut-1 RNA is exclusively found in DNA from uncultivated bacteria present in samples from the human gut.

The manA RNA motif refers to a conserved RNA structure that was identified by bioinformatics. Instances of the manA RNA motif were detected in bacteria in the genus Photobacterium and phages that infect certain kinds of cyanobacteria. However, most predicted manA RNA sequences are derived from DNA collected from uncultivated marine bacteria. Almost all manA RNAs are positioned such that they might be in the 5' untranslated regions of protein-coding genes, and therefore it was hypothesized that manA RNAs function as cis-regulatory elements. Given the relative complexity of their secondary structure, and their hypothesized cis-regulatory role, they might be riboswitches.

The Ocean-VII RNA motif is a conserved RNA structure that was discovered by bioinformatics. Ocean-VII motifs are found in metagenomic sequences isolated from various marine environments, and are not yet known in any classified organism. This environmental context is similar to other marine RNAs that were found previously by predominantly bioinformatic or experimental methods.