The gene rpoF encodes the sigma factor sigma-28, a protein in Escherichia coli and other species of bacteria. Depending on the bacterial species, this gene may be referred to as sigD or fliA. The protein encoded by this gene has been found to be necessary for flagellum formation.

The micF RNA is a non-coding RNA stress response gene found in Escherichia coli and related bacteria that post-transcriptionally controls expression of the outer membrane porin gene ompF. The micF gene encodes a non-translated 93 nucleotide antisense RNA that binds its target ompF mRNA and regulates ompF expression by inhibiting translation and inducing degradation of the message. In addition, other factors, such as the RNA chaperone protein StpA also play a role in this regulatory system. The expression of micF is controlled by both environmental and internal stress factors. Four transcriptional regulators are known to bind the micF promoter region and activate micF expression.

The repression of heat shock gene expression (ROSE) element is an RNA element found in the 5' UTR of some heat shock protein's mRNAs. The ROSE element is an RNA thermometer that negatively regulates heat shock gene expression. The secondary structure is thought to be altered by temperature, thus it is an RNA thermometer. This structure blocks access to the ribosome binding site at normal temperatures. During heat shock however, the structure changes freeing the ribosome binding site and allowing expression to occur.

The rsmY RNA family is a set of related non-coding RNA genes, that like RsmZ, is regulated by the GacS/GacA signal transduction system in the plant-beneficial soil bacterium and biocontrol model organism Pseudomonas fluorescens CHA0. GacA/GacS target genes are translationally repressed by the small RNA binding protein RsmA. RsmY and RsmZ RNAs bind RsmA to relieve this repression and so enhance secondary metabolism and biocontrol traits.

The hok/sok system is a postsegregational killing mechanism employed by the R1 plasmid in Escherichia coli. It was the first type I toxin-antitoxin pair to be identified through characterisation of a plasmid-stabilising locus. It is a type I system because the toxin is neutralised by a complementary RNA, rather than a partnered protein.
In biology, an autoinducer is a signaling molecule that enables detection and response to changes in the population density of bacterial cells. Synthesized when a bacterium reproduces, autoinducers pass outside the bacterium and into the surrounding medium. They are a key component of the phenomenon of quorum sensing: as the density of quorum-sensing bacterial cells increases, so does the concentration of the autoinducer. A bacterium’s detection of an autoinducer above some minimum threshold triggers altered gene expression.
An upstream open reading frame (uORF) is an open reading frame (ORF) within the 5' untranslated region (5'UTR) of an mRNA. uORFs can regulate eukaryotic gene expression. Translation of the uORF typically inhibits downstream expression of the primary ORF. However, in some genes such as yeast GCN4, translation of specific uORFs may increase translation of the main ORF. In bacteria, uORFs are called leader peptides and were originally discovered on the basis of their impact on the regulation of genes involved in the synthesis or transport of amino acids.

The Downstream-peptide motif refers to a conserved RNA structure identified by bioinformatics in the cyanobacterial genera Synechococcus and Prochlorococcus and one phage that infects such bacteria. It was also detected in marine samples of DNA from uncultivated bacteria, which are presumably other species of cyanobacteria.
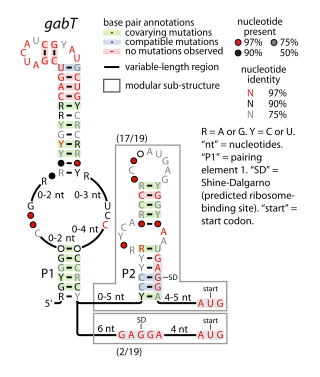
The gabT RNA motif is the name of a conserved RNA structure identified by bioinformatics whose function is unknown. The gabT motif has been detected exclusively in bacteria within the genus Pseudomonas, and is found only upstream of gabT genes, and downstream to gabD genes.

The gyrA RNA motif is a conserved RNA structure identified by bioinformatics. The RNAs are present in multiple species of bacteria within the order Pseudomonadales. This order contains the genus Pseudomonas, which includes the opportunistic human pathogen Pseudomonas aeruginosa and Pseudomonas syringae, a plant pathogen.
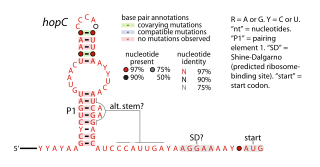
The hopC RNA motif is a predicted cis-regulatory element identified by a bioinformatic screen for conserved RNA secondary structures. hopC RNAs are exclusively found within bacteria classified within the genus Helicobacter, some of which are human pathogens that infect the stomach and can cause ulcers.

The JUMPstart RNA motif describes a conserved RNA-based secondary structure associated with JUMPstart elements. The 39-base-pair JUMPstart sequence describes a conserved element upstream of genes that participate in polysaccharide synthesis. The JUMPstart element has been shown to function as an RNA, and is present in the 5' untranslated regions of the genes it regulates.

The rmf RNA motif is a conserved RNA structure that was originally detected using bioinformatics. rmf RNAs are consistently foundwithin species classified into the genus Pseudomonas, and is located potentially in the 5′ untranslated regions of rmf genes. These genes encodes the ribosome modulation factor protein, which affects the translation of genes by modifying ribosome structure in response to stress such as starvation. This ribosome modulation is a part of the stringent response in bacteria. The likely biological role of rmf RNAs is ambiguous. Since the RNA could be in the 5′ UTRs of protein-coding genes, it was hypothesized that it functions as a cis-regulatory element. This hypothesis is bolstered by the observation that ribosome modulation factor binds ribosomal RNA, and many cis-regulatory RNAs called ribosomal protein leaders participate in a feedback regulation mechanism by binding to proteins that normally bind to ribosomal RNA. However, since rmf RNAs are not very close to the rmf genes, they might function as non-coding RNAs.

The traJ-II RNA motif is a conserved RNA structure discovered in bacteria by using bioinformatics. traJ-II RNAs appear to be in the 5' untranslated regions of protein-coding genes called traJ, which functions in the process of bacterial conjugation. A previously identified motif known as TraJ 5' UTR is also found upstream of traJ genes functions as the target of FinP antisense RNAs, so it is possible that traJ-II RNAs play a similar role as targets of an antisense RNA. However, some sequence features within the traJ-II RNA motif suggest that the biological RNA might be transcribed from the reverse-complement strand. Thus is it unclear whether traJ-II function as cis-regulatory elements. traJ-II RNAs are found in a variety of Pseudomonadota.

The rsmX gene is part of the Rsm/Csr family of non-coding RNAs (ncRNAs). Members of the Rsm/Csr family are present in a diverse range of bacteria, including Escherichia coli, Erwinia, Salmonella, Vibrio and Pseudomonas. These ncRNAs act by sequestering translational repressor proteins, called RsmA, activating expression of downstream genes that would normally be blocked by the repressors. Sequestering of target proteins is dependent upon exposed GGA motifs in the stem loops of the ncRNAs. Typically, the activated genes are involved in secondary metabolism, biofilm formation and motility.
Bacterial small RNAs are small RNAs produced by bacteria; they are 50- to 500-nucleotide non-coding RNA molecules, highly structured and containing several stem-loops. Numerous sRNAs have been identified using both computational analysis and laboratory-based techniques such as Northern blotting, microarrays and RNA-Seq in a number of bacterial species including Escherichia coli, the model pathogen Salmonella, the nitrogen-fixing alphaproteobacterium Sinorhizobium meliloti, marine cyanobacteria, Francisella tularensis, Streptococcus pyogenes, the pathogen Staphylococcus aureus, and the plant pathogen Xanthomonas oryzae pathovar oryzae. Bacterial sRNAs affect how genes are expressed within bacterial cells via interaction with mRNA or protein, and thus can affect a variety of bacterial functions like metabolism, virulence, environmental stress response, and structure.

CrcZ is a small RNA found in Pseudomonas bacteria, which acts as a global regulator of carbon catabolite repression. In P. aeruginosa, CrcZ is responsible for sequestering the protein Crc. Crc is an RNA-binding global regulator, which acts by inhibiting the translation of the transcriptional regulator AlkS.

The COG3610-DE RNA motif is a conserved RNA structure that was discovered by bioinformatics. COG3610-DE motifs are found in the genus Lactobacillales, which is part of the phylum Bacillota.

The uup RNA motif is a conserved RNA structure that was discovered by bioinformatics. uup motif RNAs are found in Bacillota and Gammaproteobacteria.















