
The centromere links a pair of sister chromatids together during cell division. This constricted region of chromosome connects the sister chromatids, creating a short arm (p) and a long arm (q) on the chromatids. During mitosis, spindle fibers attach to the centromere via the kinetochore.

Cell division is the process by which a parent cell divides into two daughter cells. Cell division usually occurs as part of a larger cell cycle in which the cell grows and replicates its chromosome(s) before dividing. In eukaryotes, there are two distinct types of cell division: a vegetative division (mitosis), producing daughter cells genetically identical to the parent cell, and a cell division that produces haploid gametes for sexual reproduction (meiosis), reducing the number of chromosomes from two of each type in the diploid parent cell to one of each type in the daughter cells. Mitosis is a part of the cell cycle, in which, replicated chromosomes are separated into two new nuclei. Cell division gives rise to genetically identical cells in which the total number of chromosomes is maintained. In general, mitosis is preceded by the S stage of interphase and is followed by telophase and cytokinesis; which divides the cytoplasm, organelles, and cell membrane of one cell into two new cells containing roughly equal shares of these cellular components. The different stages of mitosis all together define the M phase of an animal cell cycle—the division of the mother cell into two genetically identical daughter cells. To ensure proper progression through the cell cycle, DNA damage is detected and repaired at various checkpoints throughout the cycle. These checkpoints can halt progression through the cell cycle by inhibiting certain cyclin-CDK complexes. Meiosis undergoes two divisions resulting in four haploid daughter cells. Homologous chromosomes are separated in the first division of meiosis, such that each daughter cell has one copy of each chromosome. These chromosomes have already been replicated and have two sister chromatids which are then separated during the second division of meiosis. Both of these cell division cycles are used in the process of sexual reproduction at some point in their life cycle. Both are believed to be present in the last eukaryotic common ancestor.
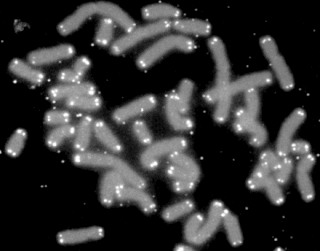
A telomere is a region of repetitive nucleotide sequences associated with specialized proteins at the ends of linear chromosomes. Telomeres are a widespread genetic feature most commonly found in eukaryotes. In most, if not all species possessing them, they protect the terminal regions of chromosomal DNA from progressive degradation and ensure the integrity of linear chromosomes by preventing DNA repair systems from mistaking the very ends of the DNA strand for a double-strand break.

Werner syndrome (WS) or Werner's syndrome, also known as "adult progeria", is a rare, autosomal recessive disorder which is characterized by the appearance of premature aging.

DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA damage, resulting in tens of thousands of individual molecular lesions per cell per day. Many of these lesions cause structural damage to the DNA molecule and can alter or eliminate the cell's ability to transcribe the gene that the affected DNA encodes. Other lesions induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells after it undergoes mitosis. As a consequence, the DNA repair process is constantly active as it responds to damage in the DNA structure. When normal repair processes fail, and when cellular apoptosis does not occur, irreparable DNA damage may occur. This can eventually lead to malignant tumors, or cancer as per the two-hit hypothesis.

S phase (Synthesis phase) is the phase of the cell cycle in which DNA is replicated, occurring between G1 phase and G2 phase. Since accurate duplication of the genome is critical to successful cell division, the processes that occur during S-phase are tightly regulated and widely conserved.

Non-homologous end joining (NHEJ) is a pathway that repairs double-strand breaks in DNA. It is called "non-homologous" because the break ends are directly ligated without the need for a homologous template, in contrast to homology directed repair (HDR), which requires a homologous sequence to guide repair. NHEJ is active in both non-dividing and proliferating cells, while HDR is not readily accessible in non-dividing cells. The term "non-homologous end joining" was coined in 1996 by Moore and Haber.

Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids.
Recombination hotspots are regions in a genome that exhibit elevated rates of recombination relative to a neutral expectation. The recombination rate within hotspots can be hundreds of times that of the surrounding region. Recombination hotspots result from higher DNA break formation in these regions, and apply to both mitotic and meiotic cells. This appellation can refer to recombination events resulting from the uneven distribution of programmed meiotic double-strand breaks.

Werner syndrome ATP-dependent helicase, also known as DNA helicase, RecQ-like type 3, is an enzyme that in humans is encoded by the WRN gene. WRN is a member of the RecQ Helicase family. Helicase enzymes generally unwind and separate double-stranded DNA. These activities are necessary before DNA can be copied in preparation for cell division. Helicase enzymes are also critical for making a blueprint of a gene for protein production, a process called transcription. Further evidence suggests that Werner protein plays a critical role in repairing DNA. Overall, this protein helps maintain the structure and integrity of a person's DNA.
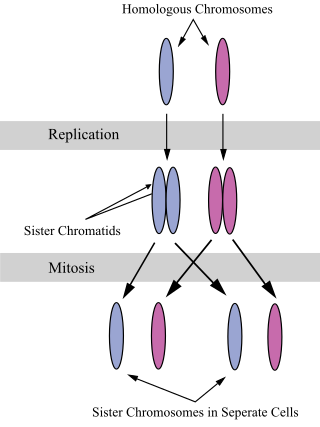
A sister chromatid refers to the identical copies (chromatids) formed by the DNA replication of a chromosome, with both copies joined together by a common centromere. In other words, a sister chromatid may also be said to be 'one-half' of the duplicated chromosome. A pair of sister chromatids is called a dyad. A full set of sister chromatids is created during the synthesis (S) phase of interphase, when all the chromosomes in a cell are replicated. The two sister chromatids are separated from each other into two different cells during mitosis or during the second division of meiosis.
Mitotic recombination is a type of genetic recombination that may occur in somatic cells during their preparation for mitosis in both sexual and asexual organisms. In asexual organisms, the study of mitotic recombination is one way to understand genetic linkage because it is the only source of recombination within an individual. Additionally, mitotic recombination can result in the expression of recessive alleles in an otherwise heterozygous individual. This expression has important implications for the study of tumorigenesis and lethal recessive alleles. Mitotic homologous recombination occurs mainly between sister chromatids subsequent to replication. Inter-sister homologous recombination is ordinarily genetically silent. During mitosis the incidence of recombination between non-sister homologous chromatids is only about 1% of that between sister chromatids.

Serine/threonine-protein kinase ATR, also known as ataxia telangiectasia and Rad3-related protein (ATR) or FRAP-related protein 1 (FRP1), is an enzyme that, in humans, is encoded by the ATR gene. It is a large kinase of about 301.66 kDa. ATR belongs to the phosphatidylinositol 3-kinase-related kinase protein family. ATR is activated in response to single strand breaks, and works with ATM to ensure genome integrity.
Chromatin remodeling is the dynamic modification of chromatin architecture to allow access of condensed genomic DNA to the regulatory transcription machinery proteins, and thereby control gene expression. Such remodeling is principally carried out by 1) covalent histone modifications by specific enzymes, e.g., histone acetyltransferases (HATs), deacetylases, methyltransferases, and kinases, and 2) ATP-dependent chromatin remodeling complexes which either move, eject or restructure nucleosomes. Besides actively regulating gene expression, dynamic remodeling of chromatin imparts an epigenetic regulatory role in several key biological processes, egg cells DNA replication and repair; apoptosis; chromosome segregation as well as development and pluripotency. Aberrations in chromatin remodeling proteins are found to be associated with human diseases, including cancer. Targeting chromatin remodeling pathways is currently evolving as a major therapeutic strategy in the treatment of several cancers.
The MRN complex is a protein complex consisting of Mre11, Rad50 and Nbs1. In eukaryotes, the MRN/X complex plays an important role in the initial processing of double-strand DNA breaks prior to repair by homologous recombination or non-homologous end joining. The MRN complex binds avidly to double-strand breaks both in vitro and in vivo and may serve to tether broken ends prior to repair by non-homologous end joining or to initiate DNA end resection prior to repair by homologous recombination. The MRN complex also participates in activating the checkpoint kinase ATM in response to DNA damage. Production of short single-strand oligonucleotides by Mre11 endonuclease activity has been implicated in ATM activation by the MRN complex.
Alternative Lengthening of Telomeres is a telomerase-independent mechanism by which cancer cells avoid the degradation of telomeres.
Chromosomal instability (CIN) is a type of genomic instability in which chromosomes are unstable, such that either whole chromosomes or parts of chromosomes are duplicated or deleted. More specifically, CIN refers to the increase in rate of addition or loss of entire chromosomes or sections of them. The unequal distribution of DNA to daughter cells upon mitosis results in a failure to maintain euploidy leading to aneuploidy. In other words, the daughter cells do not have the same number of chromosomes as the cell they originated from. Chromosomal instability is the most common form of genetic instability and cause of aneuploidy.
Telomeres, the caps on the ends of eukaryotic chromosomes, play critical roles in cellular aging and cancer. An important facet to how telomeres function in these roles is their involvement in cell cycle regulation.
The INO80 subfamily of chromatin remodeling complexes are ATPases, and includes the INO80 and SWR1 complexes.

DNA end resection, also called 5′–3′ degradation, is a biochemical process where the blunt end of a section of double-stranded DNA (dsDNA) is modified by cutting away some nucleotides from the 5' end to produce a 3' single-stranded sequence. The presence of a section of single-stranded DNA (ssDNA) allows the broken end of the DNA to line up accurately with a matching sequence, so that it can be accurately repaired.












