Related Research Articles

Plant hormones are signal molecules,produced within plants,that occur in extremely low concentrations. Plant hormones control all aspects of plant growth and development,from embryogenesis,the regulation of organ size,pathogen defense,stress tolerance and through to reproductive development. Unlike in animals each plant cell is capable of producing hormones. Went and Thimann coined the term "phytohormone" and used it in the title of their 1937 book.

Abscisic acid is a plant hormone. ABA functions in many plant developmental processes,including seed and bud dormancy,the control of organ size and stomatal closure. It is especially important for plants in the response to environmental stresses,including drought,soil salinity,cold tolerance,freezing tolerance,heat stress and heavy metal ion tolerance.

DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter,DNA methylation typically acts to repress gene transcription. In mammals,DNA methylation is essential for normal development and is associated with a number of key processes including genomic imprinting,X-chromosome inactivation,repression of transposable elements,aging,and carcinogenesis.

In epigenetics,a paramutation is an interaction between two alleles at a single locus,whereby one allele induces a heritable change in the other allele. The change may be in the pattern of DNA methylation or histone modifications. The allele inducing the change is said to be paramutagenic,while the allele that has been epigenetically altered is termed paramutable. A paramutable allele may have altered levels of gene expression,which may continue in offspring which inherit that allele,even though the paramutagenic allele may no longer be present. Through proper breeding,paramutation can result in siblings that have the same genetic sequence,but with drastically different phenotypes.
Dehydrin (DHN) is a multi-family of proteins present in plants that is produced in response to cold and drought stress. DHNs are hydrophilic,reliably thermostable,and disordered. They are stress proteins with a high number of charged amino acids that belong to the Group II Late Embryogenesis Abundant (LEA) family. DHNs are primarily found in the cytoplasm and nucleus but more recently,they have been found in other organelles,like mitochondria and chloroplasts.
RNA polymerase IV is an enzyme that synthesizes small interfering RNA (siRNA) in plants,which silence gene expression. RNAP IV belongs to a family of enzymes that catalyze the process of transcription known as RNA Polymerases,which synthesize RNA from DNA templates. Discovered via phylogenetic studies of land plants,genes of RNAP IV are thought to have resulted from multistep evolution processes that occurred in RNA Polymerase II phylogenies. Such an evolutionary pathway is supported by the fact that RNAP IV is composed of 12 protein subunits that are either similar or identical to RNA polymerase II,and is specific to plant genomes. Via its synthesis of siRNA,RNAP IV is involved in regulation of heterochromatin formation in a process known as RNA directed DNA Methylation (RdDM).

Transgenerational epigenetic inheritance is the transmission of epigenetic markers and modifications from one generation to multiple subsequent generations without altering the primary structure of DNA. Thus,the regulation of genes via epigenetic mechanisms can be heritable;the amount of transcription and proteins produced can be mitigated by inherited epigenetic changes. The less precise term "epigenetic inheritance" may cover both cell–cell and organism–organism information transfer. Although these two levels of epigenetic inheritance are equivalent in unicellular organisms,they may have distinct mechanisms and evolutionary distinctions in multicellular organisms.

N6-Methyladenosine (m6A) was originally identified and partially characterised in the 1970s,and is an abundant modification in mRNA and DNA. It is found within some viruses,and most eukaryotes including mammals,insects,plants and yeast. It is also found in tRNA,rRNA,and small nuclear RNA (snRNA) as well as several long non-coding RNA,such as Xist.
In molecular biology mir-398 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
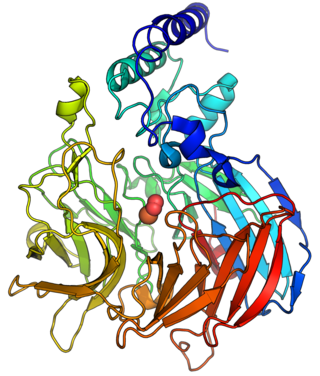
9-cis-epoxycarotenoid dioxygenase (EC 1.13.11.51,nine-cis-epoxycarotenoid dioxygenase,NCED,AtNCED3,PvNCED1,VP14) is an enzyme in the biosynthesis of abscisic acid (ABA),with systematic name 9-cis-epoxycarotenoid 11,12-dioxygenase. This enzyme catalyses the following chemical reaction

Epigenome editing or epigenome engineering is a type of genetic engineering in which the epigenome is modified at specific sites using engineered molecules targeted to those sites. Whereas gene editing involves changing the actual DNA sequence itself,epigenetic editing involves modifying and presenting DNA sequences to proteins and other DNA binding factors that influence DNA function. By "editing”epigenomic features in this manner,researchers can determine the exact biological role of an epigenetic modification at the site in question.
WRKY transcription factors are proteins that bind DNA. They are transcription factors that regulate many processes in plants and algae (Viridiplantae),such as the responses to biotic and abiotic stresses,senescence,seed dormancy and seed germination and some developmental processes but also contribute to secondary metabolism.
RNA polymerase V,previously known as RNA polymerase IVb,is a multisubunit plant specific RNA polymerase. It is required for normal function and biogenesis of small interfering RNA (siRNA). Together with RNA polymerase IV,Pol V is involved in an siRNA-dependent epigenetic pathway known as RNA-directed DNA methylation (RdDM),which establishes and maintains heterochromatic silencing in plants.
Plants depend on epigenetic processes for proper function. Epigenetics is defined as "the study of changes in gene function that are mitotically and/or meiotically heritable and that do not entail a change in DNA sequence". The area of study examines protein interactions with DNA and its associated components,including histones and various other modifications such as methylation,which alter the rate or target of transcription. Epi-alleles and epi-mutants,much like their genetic counterparts,describe changes in phenotypes due to epigenetic mechanisms. Epigenetics in plants has attracted scientific enthusiasm because of its importance in agriculture.
As a model organism,the Arabidopsis thaliana response to salinity is studied to aid understanding of other more economically important crops.

The TET enzymes are a family of ten-eleven translocation (TET) methylcytosine dioxygenases. They are instrumental in DNA demethylation. 5-Methylcytosine is a methylated form of the DNA base cytosine (C) that often regulates gene transcription and has several other functions in the genome.
Hydraulic signals in plants are detected as changes in the organism's water potential that are caused by environmental stress like drought or wounding. The cohesion and tension properties of water allow for these water potential changes to be transmitted throughout the plant.

RNA-directed DNA methylation (RdDM) is a biological process in which non-coding RNA molecules direct the addition of DNA methylation to specific DNA sequences. The RdDM pathway is unique to plants,although other mechanisms of RNA-directed chromatin modification have also been described in fungi and animals. To date,the RdDM pathway is best characterized within angiosperms,and particularly within the model plant Arabidopsis thaliana. However,conserved RdDM pathway components and associated small RNAs (sRNAs) have also been found in other groups of plants,such as gymnosperms and ferns. The RdDM pathway closely resembles other sRNA pathways,particularly the highly conserved RNAi pathway found in fungi,plants,and animals. Both the RdDM and RNAi pathways produce sRNAs and involve conserved Argonaute,Dicer and RNA-dependent RNA polymerase proteins.
Transgenerational epigenetic inheritance in plants involves mechanisms for the passing of epigenetic marks from parent to offspring that differ from those reported in animals. There are several kinds of epigenetic markers,but they all provide a mechanism to facilitate greater phenotypic plasticity by influencing the expression of genes without altering the DNA code. These modifications represent responses to environmental input and are reversible changes to gene expression patterns that can be passed down through generations. In plants,transgenerational epigenetic inheritance could potentially represent an evolutionary adaptation for sessile organisms to quickly adapt to their changing environment.

Calcium signaling in Arabidopsis is a calcium mediated signalling pathway that Arabidopsis plants use in order to respond to a stimuli. In this pathway,Ca2+ works as a long range communication ion,allowing for rapid communication throughout the plant. Systemic changes in metabolites such as glucose and sucrose takes a few minutes after the stimulus,but gene transcription occurs within seconds. Because hormones,peptides and RNA travel through the vascular system at lower speeds than the plants response to wounds,indicates that Ca2+ must be involved in the rapid signal propagation. Instead of local communication to nearby cells and tissues,Ca2+ uses mass flow within the vascular system to help with rapid transport throughout the plant. Ca2+ moving through the xylem and phloem acts through a “calcium signature”receptor system in cells where they integrate the signal and respond with the activation of defense genes. These calcium signatures encode information about the stimulus allowing the response of the plant to cater towards the type of stimulus.
References
- 1 2 3 "About Dr. Jian-Kang Zhu".
- 1 2 "Jian Kang Zhu - Google Scholar".
- ↑ "Patents by Inventor Jian-Kang Zhu".
- ↑ "Five UCR Faculty Elected AAAS Fellows".
- ↑ "UC Riverside Plant Cell Biologist Receives Top Scientific Honor".
- ↑ "王二涛研究组及合作团队提出根际微生物群落"扩增-选择"组装新模型".
- ↑ "National Science Review - Editorial Board".
- ↑ "Science China Life Sciences - Editorial Board".
- ↑ "UCR Licensed Technology Startup Companies".
- ↑ "Principal Investigators".
- ↑ Gong, Z.; Koiwa, H.; Cushman, M. A.; Ray, A.; Bufford, D.; Kore-Eda, S.; Matsumoto, T. K.; Zhu, J.; Cushman, J. C.; Bressan, R. A.; Hasegawa, P. M. (2001). "Genes That Are Uniquely Stress Regulated in Salt Overly Sensitive (sos) Mutants1". Plant Physiology. 126 (1): 363–375. doi:10.1104/pp.126.1.363. PMC 102310 . PMID 11351099.
- ↑ Xiong, Liming; Zhu, Jian-Kang (2003). "Regulation of Abscisic Acid Biosynthesis". Plant Physiology. 133 (1): 29–36. doi:10.1104/pp.103.025395. PMC 523868 . PMID 12970472.
- ↑ Zhu, J.; Lu, T.; Yue, S.; Shen, X.; Gao, F.; Busuttil, R. W.; Kupiec-Weglinski, J. W.; Xia, Q.; Zhai, Y. (2015). "Rapamycin protection of livers from ischemia and reperfusion injury is dependent on both autophagy induction and mammalian target of rapamycin complex 2-Akt activation". Transplantation. 99 (1): 48–55. doi:10.1097/TP.0000000000000476. PMC 4272660 . PMID 25340604.
- ↑ Xiong, L.; Zhu, J. K. (2003). "Regulation of Abscisic Acid Biosynthesis". Plant Physiology. 133 (1): 29–36. doi:10.1104/pp.103.025395. PMC 523868 . PMID 12970472.
- ↑ Sunkar, R.; Zhu, J. K. (2004). "Novel and Stress-Regulated MicroRNAs and Other Small RNAs from Arabidopsis". The Plant Cell. 16 (8): 2001–2019. doi:10.1105/tpc.104.022830. PMC 519194 . PMID 15258262.
- ↑ Agius, F.; Kapoor, A.; Zhu, J. K. (2006). "Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation". Proceedings of the National Academy of Sciences of the United States of America. 103 (31): 11796–11801. Bibcode:2006PNAS..10311796A. doi: 10.1073/pnas.0603563103 . PMC 1544249 . PMID 16864782.
- ↑ "A protein complex regulates RNA processing of intronic heterochromatin-containing genes in Arabidopsis".
- ↑ Huang, Huan; Liu, Ruie; Niu, Qingfeng; Tang, Kai; Zhang, Bo; Zhang, Heng; Chen, Kunsong; Zhu, Jian-Kang; Lang, Zhaobo (2019). "Global increase in DNA methylation during orange fruit development and ripening". Proceedings of the National Academy of Sciences. 116 (4): 1430–1436. Bibcode:2019PNAS..116.1430H. doi: 10.1073/pnas.1815441116 . PMC 6347674 . PMID 30635417.
- ↑ Mao, Yanfei; Yang, Xiaoxuan; Zhou, Yiting; Zhang, Zhengjing; Botella, Jose Ramon; Zhu, Jian-Kang (2018). "Manipulating plant RNA-silencing pathways to improve the gene editing efficiency of CRISPR/Cas9 systems". Genome Biology. 19 (1): 149. doi: 10.1186/s13059-018-1529-7 . PMC 6161460 . PMID 30266091.
- ↑ Miki, D.; Zhang, W.; Zeng, W.; Feng, Z.; Zhu, J. K. (2018). "CRISPR/Cas9-mediated gene targeting in Arabidopsis using sequential transformation". Nature Communications. 9 (1): 1967. Bibcode:2018NatCo...9.1967M. doi:10.1038/s41467-018-04416-0. PMC 5958078 . PMID 29773790.
- ↑ "Indiana Thinkers Make 'Most Influential Minds' List".
- ↑ "Jian Kang Zhu - 2016 Herbert Newby McCoy Award".
- ↑ "Jian-Kang Zhu".
- ↑ "ASPB Pioneer Members".