Related Research Articles

MicroRNA (miRNA) are small, single-stranded, non-coding RNA molecules containing 21 to 23 nucleotides. Found in plants, animals and some viruses, miRNAs are involved in RNA silencing and post-transcriptional regulation of gene expression. miRNAs base-pair to complementary sequences in mRNA molecules, then silence said mRNA molecules by one or more of the following processes:
- Cleavage of the mRNA strand into two pieces,
- Destabilization of the mRNA by shortening its poly(A) tail, or
- Reducing translation of the mRNA into proteins.

A non-coding RNA (ncRNA) is a functional RNA molecule that is not translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important types of non-coding RNAs include transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as small RNAs such as microRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, exRNAs, scaRNAs and the long ncRNAs such as Xist and HOTAIR.
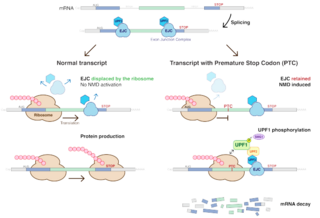
Nonsense-mediated mRNA decay (NMD) is a surveillance pathway that exists in all eukaryotes. Its main function is to reduce errors in gene expression by eliminating mRNA transcripts that contain premature stop codons. Translation of these aberrant mRNAs could, in some cases, lead to deleterious gain-of-function or dominant-negative activity of the resulting proteins.
Myc is a family of regulator genes and proto-oncogenes that code for transcription factors. The Myc family consists of three related human genes: c-myc (MYC), l-myc (MYCL), and n-myc (MYCN). c-myc was the first gene to be discovered in this family, due to homology with the viral gene v-myc.
The Let-7 microRNA precursor was identified from a study of developmental timing in C. elegans, and was later shown to be part of a much larger class of non-coding RNAs termed microRNAs. miR-98 microRNA precursor from human is a let-7 family member. Let-7 miRNAs have now been predicted or experimentally confirmed in a wide range of species (MIPF0000002). miRNAs are initially transcribed in long transcripts called primary miRNAs (pri-miRNAs), which are processed in the nucleus by Drosha and Pasha to hairpin structures of about 70 nucleotide. These precursors (pre-miRNAs) are exported to the cytoplasm by exportin5, where they are subsequently processed by the enzyme Dicer to a ~22 nucleotide mature miRNA. The involvement of Dicer in miRNA processing demonstrates a relationship with the phenomenon of RNA interference.

The miR-9 microRNA, is a short non-coding RNA gene involved in gene regulation. The mature ~21nt miRNAs are processed from hairpin precursor sequences by the Dicer enzyme. The dominant mature miRNA sequence is processed from the 5' arm of the mir-9 precursor, and from the 3' arm of the mir-79 precursor. The mature products are thought to have regulatory roles through complementarity to mRNA. In vertebrates, miR-9 is highly expressed in the brain, and is suggested to regulate neuronal differentiation. A number of specific targets of miR-9 have been proposed, including the transcription factor REST and its partner CoREST.

The miR-17 microRNA precursor family are a group of related small non-coding RNA genes called microRNAs that regulate gene expression. The microRNA precursor miR-17 family, includes miR-20a/b, miR-93, and miR-106a/b. With the exception of miR-93, these microRNAs are produced from several microRNA gene clusters, which apparently arose from a series of ancient evolutionary genetic duplication events, and also include members of the miR-19, and miR-25 families. These clusters are transcribed as long non-coding RNA transcripts that are processed to form ~70 nucleotide microRNA precursors, that are subsequently processed by the Dicer enzyme to give a ~22 nucleotide products. The mature microRNA products are thought to regulate expression levels of other genes through complementarity to the 3' UTR of specific target messenger RNA.
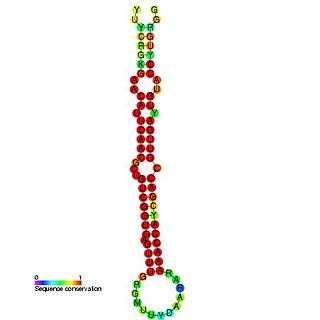
In molecular biology miR-181 microRNA precursor is a small non-coding RNA molecule. MicroRNAs (miRNAs) are transcribed as ~70 nucleotide precursors and subsequently processed by the RNase-III type enzyme Dicer to give a ~22 nucleotide mature product. In this case the mature sequence comes from the 5' arm of the precursor. They target and modulate protein expression by inhibiting translation and / or inducing degradation of target messenger RNAs. This new class of genes has recently been shown to play a central role in malignant transformation. miRNA are downregulated in many tumors and thus appear to function as tumor suppressor genes. The mature products miR-181a, miR-181b, miR-181c or miR-181d are thought to have regulatory roles at posttranscriptional level, through complementarity to target mRNAs. miR-181 has been predicted or experimentally confirmed in a wide number of vertebrate species such as rat, zebrafish, and pufferfish.

There are 89 known sequences today in the microRNA 19 (miR-19) family but it will change quickly. They are found in a large number of vertebrate species. The miR-19 microRNA precursor is a small non-coding RNA molecule that regulates gene expression. Within the human and mouse genome there are three copies of this microRNA that are processed from multiple predicted precursor hairpins:

mRNA-decapping enzyme 2 is a protein that in humans is encoded by the DCP2 gene.

Putative microRNA host gene 1 protein is a protein that in humans is encoded by the MIR17HG gene.
Post-transcriptional regulation is the control of gene expression at the RNA level. It occurs once the RNA polymerase has been attached to the gene's promoter and is synthesizing the nucleotide sequence. Therefore, as the name indicates, it occurs between the transcription phase and the translation phase of gene expression. These controls are critical for the regulation of many genes across human tissues. It also plays a big role in cell physiology, being implicated in pathologies such as cancer and neurodegenerative diseases.

MiR-155 is a microRNA that in humans is encoded by the MIR155 host gene or MIR155HG. MiR-155 plays a role in various physiological and pathological processes. Exogenous molecular control in vivo of miR-155 expression may inhibit malignant growth, viral infections, and enhance the progression of cardiovascular diseases.

In molecular biology mir-126 is a short non-coding RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several pre- and post-transcription mechanisms.

In molecular biology, mir-145 microRNA is a short RNA molecule that in humans is encoded by the MIR145 gene. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.

In molecular biology mir-22 microRNA is a short RNA molecule. MicroRNAs are an abundant class of molecules, approximately 22 nucleotides in length, which can post-transcriptionally regulate gene expression by binding to the 3' UTR of mRNAs expressed in a cell.

In molecular biology mir-210 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-339 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms. miR-339-5p expression was associated with overall survival in breast cancer.
In molecular biology mir-345 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
References
- 1 2 3 4 5 HHMI biography
- ↑ Mendell, J. T.; Medghalchi, S. M.; Lake, R. G.; Noensie, E. N.; Dietz, H. C. (2000). "Novel Upf2p Orthologues Suggest a Functional Link between Translation Initiation and Nonsense Surveillance Complexes". Molecular and Cellular Biology. 20 (23): 8944–8957. doi:10.1128/MCB.20.23.8944-8957.2000. PMC 86549 . PMID 11073994.
- ↑ Medghalchi, S. M.; Frischmeyer, P. A.; Mendell, J. T.; Kelly, A. G.; Lawler, A. M.; Dietz, H. C. (2001). "Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability". Human Molecular Genetics. 10 (2): 99–105. doi: 10.1093/hmg/10.2.99 . PMID 11152657.
- ↑ Mendell, J. T.; Dietz, H. C. (2001). "When the message goes awry: disease-producing mutations that influence mRNA content and performance". Cell. 107 (4): 411–414. doi: 10.1016/S0092-8674(01)00583-9 . PMID 11719181.
- ↑ Mendell, J.; Ap Rhys, C.; Dietz, H. (2002). "Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts". Science. 298 (5592): 419–422. doi: 10.1126/science.1074428 . PMID 12228722. S2CID 46447414.
- ↑ Mendell, J.; Sharifi, N.; Meyers, J.; Martinez-Murillo, F.; Dietz, H. (2004). "Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise". Nature Genetics. 36 (10): 1073–1078. doi: 10.1038/ng1429 . PMID 15448691.
- ↑ Dews, M.; Homayouni, A.; Yu, D.; Murphy, D.; Sevignani, C.; Wentzel, E.; Furth, E.; Lee, W.; Enders, G.; Mendell, J. T.; Thomas-Tikhonenko, A. (2006). "Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster". Nature Genetics. 38 (9): 1060–1065. doi:10.1038/ng1855. PMC 2669546 . PMID 16878133.
- ↑ Chang, T.; Yu, D.; Lee, Y.; Wentzel, E.; Arking, D.; West, K.; Dang, C.; Thomas-Tikhonenko, A.; Mendell, J. (2008). "Widespread microRNA repression by Myc contributes to tumorigenesis". Nature Genetics. 40 (1): 43–50. doi:10.1038/ng.2007.30. PMC 2628762 . PMID 18066065.
- ↑ Chang, T.; Zeitels, L.; Hwang, H.; Chivukula, R.; Wentzel, E.; Dews, M.; Jung, J.; Gao, P.; Dang, C.; Beer, M. A.; Thomas-Tikhonenko, A.; Mendell, J. T. (2009). "Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation". Proceedings of the National Academy of Sciences of the United States of America. 106 (9): 3384–3389. Bibcode:2009PNAS..106.3384C. doi: 10.1073/pnas.0808300106 . PMC 2651245 . PMID 19211792.
- ↑ Chang, T.; Wentzel, E.; Kent, O.; Ramachandran, K.; Mullendore, M.; Lee, K.; Feldmann, G.; Yamakuchi, M.; Ferlito, M.; Lowenstein, C. J.; Arking, D. E.; Beer, M. A.; Maitra, A.; Mendell, J. T. (2007). "Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis". Molecular Cell. 26 (5): 745–752. doi:10.1016/j.molcel.2007.05.010. PMC 1939978 . PMID 17540599.
- ↑ Gao, P.; Tchernyshyov, I.; Chang, T.; Lee, Y.; Kita, K.; Ochi, T.; Zeller, K.; De Marzo, A.; Van Eyk, J.; Mendell, J. T.; Dang, C. V. (2009). "c-Myc suppression of miR-23 enhances mitochondrial glutaminase and glutamine metabolism". Nature. 458 (7239): 762–765. Bibcode:2009Natur.458..762G. doi:10.1038/nature07823. PMC 2729443 . PMID 19219026.
- ↑ Hwang, H.; Wentzel, E.; Mendell, J. (2009). "Cell–cell contact globally activates microRNA biogenesis". Proceedings of the National Academy of Sciences of the United States of America. 106 (17): 7016–7021. Bibcode:2009PNAS..106.7016H. doi: 10.1073/pnas.0811523106 . PMC 2678439 . PMID 19359480.
- ↑ Chivukula, R.; Mendell, J. (2009). "Abate and switch: miR-145 in stem cell differentiation". Cell. 137 (4): 606–608. doi: 10.1016/j.cell.2009.04.059 . PMID 19450510.
- ↑ Kota, J.; Chivukula, R.; O'Donnell, K.; Wentzel, E.; Montgomery, C.; Hwang, H.; Chang, T.; Vivekanandan, P.; Torbenson, M.; Clark, K. R.; Mendell, J. R.; Mendell, J. T. (2009). "Therapeutic delivery of miR-26a inhibits cancer cell proliferation and induces tumor-specific apoptosis". Cell. 137 (6): 1005–1017. doi:10.1016/j.cell.2009.04.021. PMC 2722880 . PMID 19524505.
- ↑ Replacing microRNA for cancer treatment. Jenny Lauren Lee, Science News, July 4, 2009
- ↑ MicroRNA Replacement Therapy May Stop Cancer In Its Tracks Science Daily, June 12, 2009
- ↑ Cancer May Be Stopped In Its Tracks By MicroRNA Replacement Therapy Medical News Today, June 13, 2009
- ↑ Dorian Gray: Is there a secret to eternal youth? [ dead link ] Anjana Ahuja, The Times, September 8, 2009
- ↑ Hwang, H.; Mendell, J. (2006). "MicroRNAs in cell proliferation, cell death, and tumorigenesis". British Journal of Cancer. 94 (6): 776–780. doi:10.1038/sj.bjc.6603023. PMC 2361377 . PMID 16495913.
- ↑ Kent, O.; Mendell, J. (2006). "A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes". Oncogene. 25 (46): 6188–6196. doi:10.1038/sj.onc.1209913. PMID 17028598. S2CID 36269207.
- ↑ Mendell, J. (2008). "MiRiad roles for the miR-17-92 cluster in development and disease". Cell. 133 (2): 217–222. doi:10.1016/j.cell.2008.04.001. PMC 2732113 . PMID 18423194.
- ↑ Mendell, J. T.; Panicker, S. G.; Tsao, C. Y.; Feng, B.; Sahenk, Z.; Marzluf, G. A.; Mendell, J. R. (1998). "Novel compound heterozygous laminina2-chain gene (LAMA2) mutations in congenital muscular dystrophy". Human Mutation. 12 (2): 135. doi:10.1002/(SICI)1098-1004(1998)12:2<135::AID-HUMU10>3.0.CO;2-6. PMID 10694916.