Biostatistics is a branch of statistics that applies statistical methods to a wide range of topics in biology. It encompasses the design of biological experiments, the collection and analysis of data from those experiments and the interpretation of the results.

Bioinformatics is an interdisciplinary field of science that develops methods and software tools for understanding biological data, especially when the data sets are large and complex. Bioinformatics uses biology, chemistry, physics, computer science, data science, computer programming, information engineering, mathematics and statistics to analyze and interpret biological data. The process of analyzing and interpreting data can sometimes be referred to as computational biology, however this distinction between the two terms is often disputed. To some, the term computational biology refers to building and using models of biological systems.

Proteomics is the large-scale study of proteins. Proteins are vital macromolecules of all living organisms, with many functions such as the formation of structural fibers of muscle tissue, enzymatic digestion of food, or synthesis and replication of DNA. In addition, other kinds of proteins include antibodies that protect an organism from infection, and hormones that send important signals throughout the body.

Computational biology refers to the use of techniques in computer science, data analysis, mathematical modeling and computational simulations to understand biological systems and relationships. An intersection of computer science, biology, and data science, the field also has foundations in applied mathematics, molecular biology, cell biology, chemistry, and genetics.

A DNA microarray is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome. Each DNA spot contains picomoles of a specific DNA sequence, known as probes. These can be a short section of a gene or other DNA element that are used to hybridize a cDNA or cRNA sample under high-stringency conditions. Probe-target hybridization is usually detected and quantified by detection of fluorophore-, silver-, or chemiluminescence-labeled targets to determine relative abundance of nucleic acid sequences in the target. The original nucleic acid arrays were macro arrays approximately 9 cm × 12 cm and the first computerized image based analysis was published in 1981. It was invented by Patrick O. Brown. An example of its application is in SNPs arrays for polymorphisms in cardiovascular diseases, cancer, pathogens and GWAS analysis. It is also used for the identification of structural variations and the measurement of gene expression.

Biological databases are libraries of biological sciences, collected from scientific experiments, published literature, high-throughput experiment technology, and computational analysis. They contain information from research areas including genomics, proteomics, metabolomics, microarray gene expression, and phylogenetics. Information contained in biological databases includes gene function, structure, localization, clinical effects of mutations as well as similarities of biological sequences and structures.

Systems biology is the computational and mathematical analysis and modeling of complex biological systems. It is a biology-based interdisciplinary field of study that focuses on complex interactions within biological systems, using a holistic approach to biological research.
The transcriptome is the set of all RNA transcripts, including coding and non-coding, in an individual or a population of cells. The term can also sometimes be used to refer to all RNAs, or just mRNA, depending on the particular experiment. The term transcriptome is a portmanteau of the words transcript and genome; it is associated with the process of transcript production during the biological process of transcription.
Bioconductor is a free, open source and open development software project for the analysis and comprehension of genomic data generated by wet lab experiments in molecular biology.

In the field of molecular biology, gene expression profiling is the measurement of the activity of thousands of genes at once, to create a global picture of cellular function. These profiles can, for example, distinguish between cells that are actively dividing, or show how the cells react to a particular treatment. Many experiments of this sort measure an entire genome simultaneously, that is, every gene present in a particular cell.

Genetic analysis is the overall process of studying and researching in fields of science that involve genetics and molecular biology. There are a number of applications that are developed from this research, and these are also considered parts of the process. The base system of analysis revolves around general genetics. Basic studies include identification of genes and inherited disorders. This research has been conducted for centuries on both a large-scale physical observation basis and on a more microscopic scale. Genetic analysis can be used generally to describe methods both used in and resulting from the sciences of genetics and molecular biology, or to applications resulting from this research.
Genevestigator is an application consisting of a gene expression database and tools to analyse the data. It exists in two versions, biomedical and plant, depending on the species of the underlying microarray and RNAseq as well as single-cell RNA-sequencing data. It was started in January 2004 by scientists from ETH Zurich and is currently developed and commercialized by Nebion AG.

Oncogenomics is a sub-field of genomics that characterizes cancer-associated genes. It focuses on genomic, epigenomic and transcript alterations in cancer.

Microarray analysis techniques are used in interpreting the data generated from experiments on DNA, RNA, and protein microarrays, which allow researchers to investigate the expression state of a large number of genes – in many cases, an organism's entire genome – in a single experiment. Such experiments can generate very large amounts of data, allowing researchers to assess the overall state of a cell or organism. Data in such large quantities is difficult – if not impossible – to analyze without the help of computer programs.
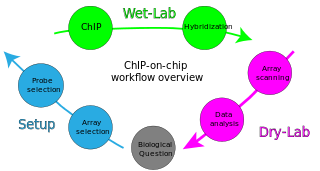
ChIP-on-chip is a technology that combines chromatin immunoprecipitation ('ChIP') with DNA microarray ("chip"). Like regular ChIP, ChIP-on-chip is used to investigate interactions between proteins and DNA in vivo. Specifically, it allows the identification of the cistrome, the sum of binding sites, for DNA-binding proteins on a genome-wide basis. Whole-genome analysis can be performed to determine the locations of binding sites for almost any protein of interest. As the name of the technique suggests, such proteins are generally those operating in the context of chromatin. The most prominent representatives of this class are transcription factors, replication-related proteins, like origin recognition complex protein (ORC), histones, their variants, and histone modifications.
A microarray database is a repository containing microarray gene expression data. The key uses of a microarray database are to store the measurement data, manage a searchable index, and make the data available to other applications for analysis and interpretation.
Geniom RT Analyzer is an instrument used in molecular biology for diagnostic testing. The Geniom RT Analyzer utilizes the dynamic nature of tissue microRNA levels as a biomarker for disease progression. The Geniom analyzer incorporates microfluidic and biochip microarray technology in order to quantify microRNAs via a Microfluidic Primer Extension Assay (MPEA) technique.
BioMart is a community-driven project to provide a single point of access to distributed research data. The BioMart project contributes open source software and data services to the international scientific community. Although the BioMart software is primarily used by the biomedical research community, it is designed in such a way that any type of data can be incorporated into the BioMart framework. The BioMart project originated at the European Bioinformatics Institute as a data management solution for the Human Genome Project. Since then, BioMart has grown to become a multi-institute collaboration involving various database projects on five continents.
Secretomics is a type of proteomics which involves the analysis of the secretome—all the secreted proteins of a cell, tissue or organism. Secreted proteins are involved in a variety of physiological processes, including cell signaling and matrix remodeling, but are also integral to invasion and metastasis of malignant cells. Secretomics has thus been especially important in the discovery of biomarkers for cancer and understanding molecular basis of pathogenesis. The analysis of the insoluble fraction of the secretome has been termed matrisomics.











