Related Research Articles

Magnetic resonance imaging (MRI) is a medical imaging technique used in radiology to form pictures of the anatomy and the physiological processes of the body. MRI scanners use strong magnetic fields, magnetic field gradients, and radio waves to generate images of the organs in the body. MRI does not involve X-rays or the use of ionizing radiation, which distinguishes it from computed tomography (CT) and positron emission tomography (PET) scans. MRI is a medical application of nuclear magnetic resonance (NMR) which can also be used for imaging in other NMR applications, such as NMR spectroscopy.

Diffusion-weighted magnetic resonance imaging is the use of specific MRI sequences as well as software that generates images from the resulting data that uses the diffusion of water molecules to generate contrast in MR images. It allows the mapping of the diffusion process of molecules, mainly water, in biological tissues, in vivo and non-invasively. Molecular diffusion in tissues is not random, but reflects interactions with many obstacles, such as macromolecules, fibers, and membranes. Water molecule diffusion patterns can therefore reveal microscopic details about tissue architecture, either normal or in a diseased state. A special kind of DWI, diffusion tensor imaging (DTI), has been used extensively to map white matter tractography in the brain.
In medicine, a biomarker is a measurable indicator of the severity or presence of some disease state. It may be defined as a "cellular, biochemical or molecular alteration in cells, tissues or fluids that can be measured and evaluated to indicate normal biological processes, pathogenic processes, or pharmacological responses to a therapeutic intervention." More generally a biomarker is anything that can be used as an indicator of a particular disease state or some other physiological state of an organism. According to the WHO, the indicator may be chemical, physical, or biological in nature - and the measurement may be functional, physiological, biochemical, cellular, or molecular.
Magnetic resonance elastography (MRE) is a form of elastography that specifically leverages MRI to quantify and subsequently map the mechanical properties of soft tissue. First developed and described at Mayo Clinic by Muthupillai et al. in 1995, MRE has emerged as a powerful, non-invasive diagnostic tool, namely as an alternative to biopsy and serum tests for staging liver fibrosis.
MRI contrast agents are contrast agents used to improve the visibility of internal body structures in magnetic resonance imaging (MRI). The most commonly used compounds for contrast enhancement are gadolinium-based contrast agents (GBCAs). Such MRI contrast agents shorten the relaxation times of nuclei within body tissues following oral or intravenous administration.
In vivo magnetic resonance spectroscopy (MRS) is a specialized technique associated with magnetic resonance imaging (MRI).
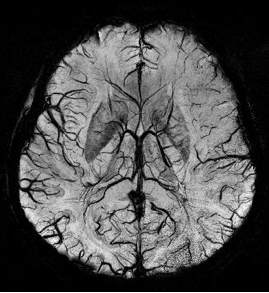
Susceptibility weighted imaging (SWI), originally called BOLD venographic imaging, is an MRI sequence that is exquisitely sensitive to venous blood, hemorrhage and iron storage. SWI uses a fully flow compensated, long echo, gradient recalled echo (GRE) pulse sequence to acquire images. This method exploits the susceptibility differences between tissues and uses the phase image to detect these differences. The magnitude and phase data are combined to produce an enhanced contrast magnitude image. The imaging of venous blood with SWI is a blood-oxygen-level dependent (BOLD) technique which is why it was referred to as BOLD venography. Due to its sensitivity to venous blood SWI is commonly used in traumatic brain injuries (TBI) and for high resolution brain venographies but has many other clinical applications. SWI is offered as a clinical package by Philips and Siemens but can be run on any manufacturer’s machine at field strengths of 1.0 T, 1.5 T, 3.0 T and higher.

Real-time magnetic resonance imaging (RT-MRI) refers to the continuous monitoring ("filming") of moving objects in real time. Because MRI is based on time-consuming scanning of k-space, real-time MRI was possible only with low image quality or low temporal resolution. Using an iterative reconstruction algorithm these limitations have recently been removed: a new method for real-time MRI achieves a temporal resolution of 20 to 30 milliseconds for images with an in-plane resolution of 1.5 to 2.0 mm. Real-time MRI promises to add important information about diseases of the joints and the heart. In many cases MRI examinations may become easier and more comfortable for patients.
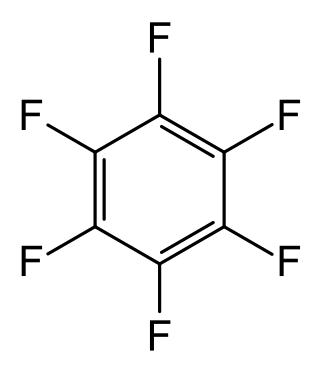
Hexafluorobenzene, HFB, C
6F
6, or perfluorobenzene is an organofluorine compound. In this derivative of benzene, all hydrogen atoms have been replaced by fluorine atoms. The technical uses of the compound are limited, although it has some specialized uses in the laboratory owing to distinctive spectroscopic properties.

Quantitative Susceptibility Mapping (QSM) provides a novel contrast mechanism in Magnetic Resonance Imaging (MRI) different from traditional Susceptibility Weighted Imaging. The voxel intensity in QSM is linearly proportional to the underlying tissue apparent magnetic susceptibility, which is useful for chemical identification and quantification of specific biomarkers including iron, calcium, gadolinium, and super paramagnetic iron oxide (SPIO) nano-particles. QSM utilizes phase images, solves the magnetic field to susceptibility source inverse problem, and generates a three-dimensional susceptibility distribution. Due to its quantitative nature and sensitivity to certain kinds of material, potential QSM applications include standardized quantitative stratification of cerebral microbleeds and neurodegenerative disease, accurate gadolinium quantification in contrast enhanced MRI, and direct monitoring of targeted theranostic drug biodistribution in nanomedicine.

Positron emission tomography–magnetic resonance imaging (PET–MRI) is a hybrid imaging technology that incorporates magnetic resonance imaging (MRI) soft tissue morphological imaging and positron emission tomography (PET) functional imaging.

Magnetic resonance imaging of the brain uses magnetic resonance imaging (MRI) to produce high quality two-dimensional or three-dimensional images of the brain and brainstem as well as the cerebellum without the use of ionizing radiation (X-rays) or radioactive tracers.
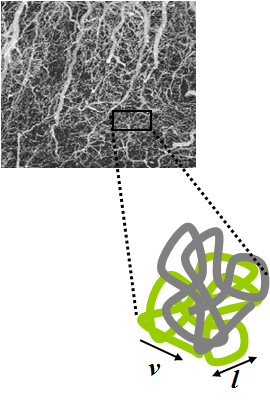
Intravoxel incoherent motion (IVIM) imaging is a concept and a method initially introduced and developed by Le Bihan et al. to quantitatively assess all the microscopic translational motions that could contribute to the signal acquired with diffusion MRI. In this model, biological tissue contains two distinct environments: molecular diffusion of water in the tissue, and microcirculation of blood in the capillary network (perfusion). The concept introduced by D. Le Bihan is that water flowing in capillaries mimics a random walk (Fig.1), as long as the assumption that all directions are represented in the capillaries is satisfied.
Functional magnetic resonance spectroscopy of the brain (fMRS) uses magnetic resonance imaging (MRI) to study brain metabolism during brain activation. The data generated by fMRS usually shows spectra of resonances, instead of a brain image, as with MRI. The area under peaks in the spectrum represents relative concentrations of metabolites.
Sodium MRI is a specialised magnetic resonance imaging technique that uses strong magnetic fields, magnetic field gradients, and radio waves to generate images of the distribution of sodium in the body, as opposed to more common forms of MRI that utilise protons present in water (1H-MRI). Like the proton, sodium is naturally abundant in the body, so can be imaged directly without the need for contrast agents or hyperpolarization. Furthermore, sodium ions play a role in important biological processes via their contribution to concentration and electrochemical gradients across cellular membranes, making it of interest as an imaging target in health and disease.
Hyperpolarized carbon-13 MRI is a functional medical imaging technique for probing perfusion and metabolism using injected substrates.
The history of magnetic resonance imaging (MRI) includes the work of many researchers who contributed to the discovery of nuclear magnetic resonance (NMR) and described the underlying physics of magnetic resonance imaging, starting early in the twentieth century. MR imaging was invented by Paul C. Lauterbur who developed a mechanism to encode spatial information into an NMR signal using magnetic field gradients in September 1971; he published the theory behind it in March 1973. The factors leading to image contrast had been described nearly 20 years earlier by physician and scientist Erik Odeblad and Gunnar Lindström. Among many other researchers in the late 1970s and 1980s, Peter Mansfield further refined the techniques used in MR image acquisition and processing, and in 2003 he and Lauterbur were awarded the Nobel Prize in Physiology or Medicine for their contributions to the development of MRI. The first clinical MRI scanners were installed in the early 1980s and significant development of the technology followed in the decades since, leading to its widespread use in medicine today.
Arterial spin labeling (ASL), also known as arterial spin tagging, is a magnetic resonance imaging technique used to quantify cerebral blood perfusion by labelling blood water as it flows throughout the brain. ASL specifically refers to magnetic labeling of arterial blood below or in the imaging slab, without the need of gadolinium contrast. A number of ASL schemes are possible, the simplest being flow alternating inversion recovery (FAIR) which requires two acquisitions of identical parameters with the exception of the out-of-slice saturation; the difference in the two images is theoretically only from inflowing spins, and may be considered a 'perfusion map'. The ASL technique was developed by Alan P. Koretsky, Donald S. Williams, John A. Detre and John S. Leigh, Jr in 1992.

Rolf Gruetter is a Swiss physicist and neurobiologist specialized in magnetic resonance, biomedical imaging and brain metabolism. He is a professor of physics at EPFL and the head of the Laboratory Functional and Metabolic Imaging at the School of Basic Sciences.
Michael Albert Thomas is an Indian-American physicist, academic, and clinical researcher. He is a Professor-in-Residence of Radiological Sciences, and Psychiatry at the Geffen School of Medicine, University of California, Los Angeles (UCLA). He is most known for developing novel single voxel based 2D NMR techniques, multi-voxel 2D MRS techniques using hybrid Cartesian as well as non-Cartesian spatio-temporal encoding such as concentric ring, radial and rosette trajectories.
References
- Magnetic resonance spectroscopic imaging entry in the public domain NCI Dictionary of Cancer Terms