
An oncogene is a gene that has the potential to cause cancer. In tumor cells, these genes are often mutated, or expressed at high levels.

A biopsy is a medical test commonly performed by a surgeon, interventional radiologist, or an interventional cardiologist. The process involves extraction of sample cells or tissues for examination to determine the presence or extent of a disease. The tissue is then fixed, dehydrated, embedded, sectioned, stained and mounted before it is generally examined under a microscope by a pathologist; it may also be analyzed chemically. When an entire lump or suspicious area is removed, the procedure is called an excisional biopsy. An incisional biopsy or core biopsy samples a portion of the abnormal tissue without attempting to remove the entire lesion or tumor. When a sample of tissue or fluid is removed with a needle in such a way that cells are removed without preserving the histological architecture of the tissue cells, the procedure is called a needle aspiration biopsy. Biopsies are most commonly performed for insight into possible cancerous or inflammatory conditions.

A neoplasm is a type of abnormal and excessive growth of tissue. The process that occurs to form or produce a neoplasm is called neoplasia. The growth of a neoplasm is uncoordinated with that of the normal surrounding tissue, and persists in growing abnormally, even if the original trigger is removed. This abnormal growth usually forms a mass, when it may be called a tumour or tumor.
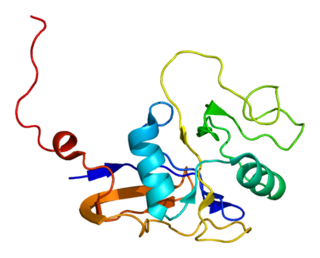
GTPase HRas, from "Harvey Rat sarcoma virus", also known as transforming protein p21 is an enzyme that in humans is encoded by the HRAS gene. The HRAS gene is located on the short (p) arm of chromosome 11 at position 15.5, from base pair 522,241 to base pair 525,549. HRas is a small G protein in the Ras subfamily of the Ras superfamily of small GTPases. Once bound to Guanosine triphosphate, H-Ras will activate a Raf kinase like c-Raf, the next step in the MAPK/ERK pathway.

KRAS is a gene that provides instructions for making a protein called K-Ras, a part of the RAS/MAPK pathway. The protein relays signals from outside the cell to the cell's nucleus. These signals instruct the cell to grow and divide (proliferate) or to mature and take on specialized functions (differentiate). It is called KRAS because it was first identified as a viral oncogene in the KirstenRAt Sarcoma virus. The oncogene identified was derived from a cellular genome, so KRAS, when found in a cellular genome, is called a proto-oncogene.
Digital polymerase chain reaction is a biotechnological refinement of conventional polymerase chain reaction methods that can be used to directly quantify and clonally amplify nucleic acids strands including DNA, cDNA, or RNA. The key difference between dPCR and traditional PCR lies in the method of measuring nucleic acids amounts, with the former being a more precise method than PCR, though also more prone to error in the hands of inexperienced users. A "digital" measurement quantitatively and discretely measures a certain variable, whereas an “analog” measurement extrapolates certain measurements based on measured patterns. PCR carries out one reaction per single sample. dPCR also carries out a single reaction within a sample, however the sample is separated into a large number of partitions and the reaction is carried out in each partition individually. This separation allows a more reliable collection and sensitive measurement of nucleic acid amounts. The method has been demonstrated as useful for studying variations in gene sequences — such as copy number variants and point mutations — and it is routinely used for clonal amplification of samples for next-generation sequencing.

Microvesicles are a type of extracellular vesicle (EV) that are released from the cell membrane. In multicellular organisms, microvesicles and other EVs are found both in tissues and in many types of body fluids. Delimited by a phospholipid bilayer, microvesicles can be as small as the smallest EVs or as large as 1000 nm. They are considered to be larger, on average, than intracellularly-generated EVs known as exosomes. Microvesicles play a role in intercellular communication and can transport molecules such as mRNA, miRNA, and proteins between cells.

Insulin-like growth factor-binding protein 3, also known as IGFBP-3, is a protein that in humans is encoded by the IGFBP3 gene. IGFBP-3 is one of six IGF binding proteins that have highly conserved structures and bind the insulin-like growth factors IGF-1 and IGF-2 with high affinity. IGFBP-7, sometimes included in this family, shares neither the conserved structural features nor the high IGF affinity. Instead, IGFBP-7 binds IGF1R, which blocks IGF-1 and IGF-2 binding, resulting in apoptosis.

DNA excision repair protein ERCC-6 is a protein that in humans is encoded by the ERCC6 gene. The ERCC6 gene is located on the long arm of chromosome 10 at position 11.23.
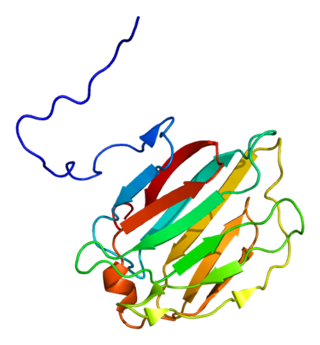
Galectin-4 is a protein that in humans is encoded by the LGALS4 gene.
Plasma cell dyscrasias are a spectrum of progressively more severe monoclonal gammopathies in which a clone or multiple clones of pre-malignant or malignant plasma cells over-produce and secrete into the blood stream a myeloma protein, i.e. an abnormal monoclonal antibody or portion thereof. The exception to this rule is the disorder termed non-secretory multiple myeloma; this disorder is a form of plasma cell dyscrasia in which no myeloma protein is detected in serum or urine of individuals who have clear evidence of an increase in clonal bone marrow plasma cells and/or evidence of clonal plasma cell-mediated tissue injury. Here, a clone of plasma cells refers to group of plasma cells that are abnormal in that they have an identical genetic identity and therefore are descendants of a single genetically distinct ancestor cell.
Recombinant adeno-associated virus (rAAV) based genome engineering is a genome editing platform centered on the use of recombinant AAV vectors that enables insertion, deletion or substitution of DNA sequences into the genomes of live mammalian cells. The technique builds on Mario Capecchi and Oliver Smithies' Nobel Prize–winning discovery that homologous recombination (HR), a natural hi-fidelity DNA repair mechanism, can be harnessed to perform precise genome alterations in mice. rAAV mediated genome-editing improves the efficiency of this technique to permit genome engineering in any pre-established and differentiated human cell line, which, in contrast to mouse ES cells, have low rates of HR.
Cell-free fetal DNA (cffDNA) is fetal DNA that circulates freely in the maternal blood. Maternal blood is sampled by venipuncture. Analysis of cffDNA is a method of non-invasive prenatal diagnosis frequently ordered for pregnant women of advanced maternal age. Two hours after delivery, cffDNA is no longer detectable in maternal blood.

Circulating tumor DNA (ctDNA) is tumor-derived fragmented DNA in the bloodstream that is not associated with cells. ctDNA should not be confused with cell-free DNA (cfDNA), a broader term which describes DNA that is freely circulating in the bloodstream, but is not necessarily of tumor origin. Because ctDNA may reflect the entire tumor genome, it has gained traction for its potential clinical utility; "liquid biopsies" in the form of blood draws may be taken at various time points to monitor tumor progression throughout the treatment regimen.
A liquid biopsy, also known as fluid biopsy or fluid phase biopsy, is the sampling and analysis of non-solid biological tissue, primarily blood. Like traditional biopsy, this type of technique is mainly used as a diagnostic and monitoring tool for diseases such as cancer, with the added benefit of being largely non-invasive. Liquid biopsies may also be used to validate the efficiency of a cancer treatment drug by taking multiple samples in the span of a few weeks. The technology may also prove beneficial for patients after treatment to monitor relapse.
Circulating free DNA (cfDNA) (also known as cell-free DNA) are degraded DNA fragments released to body fluids such as blood plasma, urine, cerebrospinal fluid, etc. Typical sizes of cfDNA fragments reflect chromatosome particles (~165bp), as well as multiples of nucleosomes, which protect DNA from digestion by apoptotic nucleases. The term cfDNA can be used to describe various forms of DNA freely circulating in body fluids, including circulating tumor DNA (ctDNA), cell-free mitochondrial DNA (ccf mtDNA), cell-free fetal DNA (cffDNA) and donor-derived cell-free DNA (dd-cfDNA). Elevated levels of cfDNA are observed in cancer, especially in advanced disease. There is evidence that cfDNA becomes increasingly frequent in circulation with the onset of age. cfDNA has been shown to be a useful biomarker for a multitude of ailments other than cancer and fetal medicine. This includes but is not limited to trauma, sepsis, aseptic inflammation, myocardial infarction, stroke, transplantation, diabetes, and sickle cell disease. cfDNA is mostly a double-stranded extracellular molecule of DNA, consisting of small fragments (50 to 200 bp) and larger fragments (21 kb) and has been recognized as an accurate marker for the diagnosis of prostate cancer and breast cancer.
Circulating mitochondrial DNA, also called cell-free circulating mitochondrial DNA and circulating cell-free mitochondrial DNA(ccf mtDNA), are short sections of mitochondrial DNA (mtDNA) that are released by cells undergoing stress or other damaging or pathological events. Circulating mitochondrial DNA is recognized by the immune system and activates inflammatory reactions. It is also a biomarker that can be used to detect the degree of damage from myocardial infarctions, cancers and ordinary stress. In certain situations it acts as a hormone.
Urinary cell-free DNA (ucfDNA) refers to DNA fragments in urine released by urogenital and non-urogenital cells. Shed cells on urogenital tract release high- or low-molecular-weight DNA fragments via apoptosis and necrosis, while circulating cell-free DNA (cfDNA) that passes through glomerular pores contributes to low-molecular-weight DNA. Most of the ucfDNA is low-molecular-weight DNA in the size of 150-250 base pairs. The detection of ucfDNA composition allows the quantification of cfDNA, circulating tumour DNA, and cell-free fetal DNA components. Many commercial kits and devices have been developed for ucfDNA isolation, quantification, and quality assessment.
Nitzan Rosenfeld is a professor of Cancer Diagnostics at the University of Cambridge. He is a Senior Group Leader at the Cancer Research UK Cambridge Institute and co-founder of Inivata, a clinical cancer genomics company.
Alain Thierry is a French geneticist and cancer researcher. He specializes in the clinical applications of circulating DNA analysis, notably in cancer care management. He is currently Director of Research at the INSERM’s Cancer Research Institute in Montpellier, France.










