
In computer programming, a string is traditionally a sequence of characters, either as a literal constant or as some kind of variable. The latter may allow its elements to be mutated and the length changed, or it may be fixed. A string is generally considered as a data type and is often implemented as an array data structure of bytes that stores a sequence of elements, typically characters, using some character encoding. String may also denote more general arrays or other sequence data types and structures.
In computer science, string-searching algorithms, sometimes called string-matching algorithms, are an important class of string algorithms that try to find a place where one or several strings are found within a larger string or text.
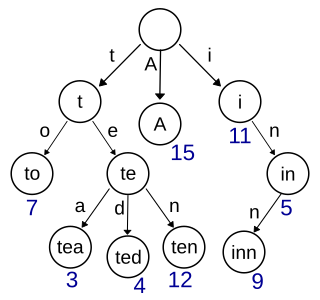
In computer science, a trie, also called digital tree or prefix tree, is a type of k-ary search tree, a tree data structure used for locating specific keys from within a set. These keys are most often strings, with links between nodes defined not by the entire key, but by individual characters. In order to access a key, the trie is traversed depth-first, following the links between nodes, which represent each character in the key.
The Burrows–Wheeler transform rearranges a character string into runs of similar characters. This is useful for compression, since it tends to be easy to compress a string that has runs of repeated characters by techniques such as move-to-front transform and run-length encoding. More importantly, the transformation is reversible, without needing to store any additional data except the position of the first original character. The BWT is thus a "free" method of improving the efficiency of text compression algorithms, costing only some extra computation. The Burrows–Wheeler transform is an algorithm used to prepare data for use with data compression techniques such as bzip2. It was invented by Michael Burrows and David Wheeler in 1994 while Burrows was working at DEC Systems Research Center in Palo Alto, California. It is based on a previously unpublished transformation discovered by Wheeler in 1983. The algorithm can be implemented efficiently using a suffix array thus reaching linear time complexity.
In computer science, the Knuth–Morris–Pratt string-searching algorithm searches for occurrences of a "word" W within a main "text string" S by employing the observation that when a mismatch occurs, the word itself embodies sufficient information to determine where the next match could begin, thus bypassing re-examination of previously matched characters.

In computer science, a suffix tree is a compressed trie containing all the suffixes of the given text as their keys and positions in the text as their values. Suffix trees allow particularly fast implementations of many important string operations.
In computer science, a suffix array is a sorted array of all suffixes of a string. It is a data structure used in, among others, full-text indices, data-compression algorithms, and the field of bibliometrics.

Zvi Galil is an Israeli-American computer scientist and mathematician. Galil served as the President of Tel Aviv University from 2007 through 2009. From 2010 to 2019, he was the dean of the Georgia Institute of Technology College of Computing. His research interests include the design and analysis of algorithms, computational complexity and cryptography. He has been credited with coining the terms stringology and sparsification. He has published over 200 scientific papers and is listed as an ISI highly cited researcher.
In computer science, the Apostolico–Giancarlo algorithm is a variant of the Boyer–Moore string-search algorithm, the basic application of which is searching for occurrences of a pattern in a text . As with other comparison-based string searches, this is done by aligning to a certain index of and checking whether a match occurs at that index. is then shifted relative to according to the rules of the Boyer–Moore algorithm, and the process repeats until the end of has been reached. Application of the Boyer-Moore shift rules often results in large chunks of the text being skipped entirely.
In computer science, compressed pattern matching is the process of searching for patterns in compressed data with little or no decompression. Searching in a compressed string is faster than searching an uncompressed string and requires less space.
In computer science, the longest palindromic substring or longest symmetric factor problem is the problem of finding a maximum-length contiguous substring of a given string that is also a palindrome. For example, the longest palindromic substring of "bananas" is "anana". The longest palindromic substring is not guaranteed to be unique; for example, in the string "abracadabra", there is no palindromic substring with length greater than three, but there are two palindromic substrings with length three, namely, "aca" and "ada". In some applications it may be necessary to return all maximal palindromic substrings rather than returning only one substring or returning the maximum length of a palindromic substring.
A factor oracle is a finite state automaton that can efficiently search for factors (substrings) in a body of text. Older techniques, such as suffix trees, were time-efficient but required significant amounts of memory. Factor oracles, by contrast, can be constructed in linear time and space in an incremental fashion.
In computer science, the longest common prefix array is an auxiliary data structure to the suffix array. It stores the lengths of the longest common prefixes (LCPs) between all pairs of consecutive suffixes in a sorted suffix array.
Wojciech Rytter is a Polish computer scientist, a professor of computer science in the automata theory group at the University of Warsaw. His research focuses on the design and analysis of algorithms, and in particular on stringology, the study of algorithms for searching and manipulating text.

In computer science, a suffix automaton is an efficient data structure for representing the substring index of a given string which allows the storage, processing, and retrieval of compressed information about all its substrings. The suffix automaton of a string is the smallest directed acyclic graph with a dedicated initial vertex and a set of "final" vertices, such that paths from the initial vertex to final vertices represent the suffixes of the string.

Gad Menahem Landau is an Israeli computer scientist noted for his contributions to combinatorial pattern matching and string algorithms and is the founding department chair of the Computer Science Department at the University of Haifa.
Gonzalo Navarro Badino is a full professor of computer science at the University of Chile and ACM Distinguished Member, whose interests include algorithms and data structures, data compression and text searching. He also participates in the Center for Biotechnology and Bioengineering and the Millennium Institute for Foundational Research on Data .. He obtained his PhD at the University of Chile in 1998 under the supervision of Ricardo Baeza-Yates with the thesis Approximate Text Searching, then worked as a post-doctoral researcher with Esko Ukkonen and Maxime Crochemore.
In computer science, the two-way string-matching algorithm is a string-searching algorithm, discovered by Maxime Crochemore and Dominique Perrin in 1991. It takes a pattern of size m, called a “needle”, preprocesses it in linear time O(m), producing information that can then be used to search for the needle in any “haystack” string, taking only linear time O(n) with n the haystack's length.
Jewels of Stringology: Text Algorithms is a book on algorithms for pattern matching in strings and related problems. It was written by Maxime Crochemore and Wojciech Rytter, and published by World Scientific in 2003.
In computer science, a generalized suffix array (or GSA) is a suffix array containing all suffixes for a set of strings. Given the set of strings of total length , it is a lexicographically sorted array of all suffixes of each string in . It is primarily used in bioinformatics and string processing.





