
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earlier along the protein sequence.

In molecular biology, the collagen triple helix or type-2 helix is the main secondary structure of various types of fibrous collagen, including type I collagen. In 1954, Ramachandran & Kartha advanced a structure for the collagen triple helix on the basis of fiber diffraction data. It consists of a triple helix made of the repetitious amino acid sequence glycine-X-Y, where X and Y are frequently proline or hydroxyproline. Collagen folded into a triple helix is known as tropocollagen. Collagen triple helices are often bundled into fibrils which themselves form larger fibres, as in tendons.

Protein secondary structure is the local spatial conformation of the polypeptide backbone excluding the side chains. The two most common secondary structural elements are alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein folds into its three dimensional tertiary structure.

Protein structure prediction is the inference of the three-dimensional structure of a protein from its amino acid sequence—that is, the prediction of its secondary and tertiary structure from primary structure. Structure prediction is different from the inverse problem of protein design. Protein structure prediction is one of the most important goals pursued by computational biology; and it is important in medicine and biotechnology.
In a chain-like biological molecule, such as a protein or nucleic acid, a structural motif is a common three-dimensional structure that appears in a variety of different, evolutionarily unrelated molecules. A structural motif does not have to be associated with a sequence motif; it can be represented by different and completely unrelated sequences in different proteins or RNA.
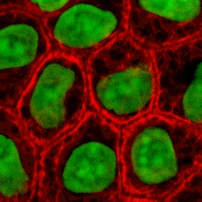
Cytokeratins are keratin proteins found in the intracytoplasmic cytoskeleton of epithelial tissue. They are an important component of intermediate filaments, which help cells resist mechanical stress. Expression of these cytokeratins within epithelial cells is largely specific to particular organs or tissues. Thus they are used clinically to identify the cell of origin of various human tumors.

Cyclic nucleotide–gated ion channels or CNG channels are ion channels that function in response to the binding of cyclic nucleotides. CNG channels are nonselective cation channels that are found in the membranes of various tissue and cell types, and are significant in sensory transduction as well as cellular development. Their function can be the result of a combination of the binding of cyclic nucleotides and either a depolarization or a hyperpolarization event. Initially discovered in the cells that make up the retina of the eye, CNG channels have been found in many different cell types across both the animal and the plant kingdoms. CNG channels have a very complex structure with various subunits and domains that play a critical role in their function. CNG channels are significant in the function of various sensory pathways including vision and olfaction, as well as in other key cellular functions such as hormone release and chemotaxis. CNG channels have also been found to exist in prokaryotes, including many spirochaeta, though their precise role in bacterial physiology remains unknown.

Mitochondrial 5-demethoxyubiquinone hydroxylase, also known as coenzyme Q7, hydroxylase, is an enzyme that in humans is encoded by the COQ7 gene. The clk-1 (clock-1) gene encodes this protein that is necessary for ubiquinone biosynthesis in the worm Caenorhabditis elegans and other eukaryotes. The mouse version of the gene is called mclk-1 and the human, fruit fly and yeast homolog COQ7.

A basic helix–loop–helix (bHLH) is a protein structural motif that characterizes one of the largest families of dimerizing transcription factors. The word "basic" does not refer to complexity but to the chemistry of the motif because transcription factors in general contain basic amino acid residues in order to facilitate DNA binding.

Major sperm protein (MSP) is a nematode specific small protein of 126 amino acids with a molecular weight of 14 kDa. It is the key player in the motility machinery of nematodes that propels the crawling movement/motility of nematode sperm. It is the most abundant protein present in nematode sperm, comprising 15% of the total protein and more than 40% of the soluble protein. MSP is exclusively synthesized in spermatocytes of the nematodes. The MSP has two main functions in the reproduction of the helminthes: i) as cytosolic component it is responsible for the crawling movement of the mature sperm, and ii) once released, it acts as hormone on the female germ cells, where it triggers oocyte maturation and stimulates the oviduct wall to contract to bring the oocytes into position for fertilization. MSP has first been identified in Caenorhabditis elegans.
A polyproline helix is a type of protein secondary structure which occurs in proteins comprising repeating proline residues. A left-handed polyproline II helix is formed when sequential residues all adopt (φ,ψ) backbone dihedral angles of roughly and have trans isomers of their peptide bonds. This PPII conformation is also common in proteins and polypeptides with other amino acids apart from proline. Similarly, a more compact right-handed polyproline I helix is formed when sequential residues all adopt (φ,ψ) backbone dihedral angles of roughly and have cis isomers of their peptide bonds. Of the twenty common naturally occurring amino acids, only proline is likely to adopt the cis isomer of the peptide bond, specifically the X-Pro peptide bond; steric and electronic factors heavily favor the trans isomer in most other peptide bonds. However, peptide bonds that replace proline with another N-substituted amino acid are also likely to adopt the cis isomer.

A pi helix is a type of secondary structure found in proteins. Discovered by crystallographer Barbara Low in 1952 and once thought to be rare, short π-helices are found in 15% of known protein structures and are believed to be an evolutionary adaptation derived by the insertion of a single amino acid into an α-helix. Because such insertions are highly destabilizing, the formation of π-helices would tend to be selected against unless it provided some functional advantage to the protein. π-helices therefore are typically found near functional sites of proteins.

A 310 helix is a type of secondary structure found in proteins and polypeptides. Of the numerous protein secondary structures present, the 310-helix is the fourth most common type observed; following α-helices, β-sheets and reverse turns. 310-helices constitute nearly 10–15% of all helices in protein secondary structures, and are typically observed as extensions of α-helices found at either their N- or C- termini. Because of the α-helices tendency to consistently fold and unfold, it has been proposed that the 310-helix serves as an intermediary conformation of sorts, and provides insight into the initiation of α-helix folding.
A helix bundle is a small protein fold composed of several alpha helices that are usually nearly parallel or antiparallel to each other.
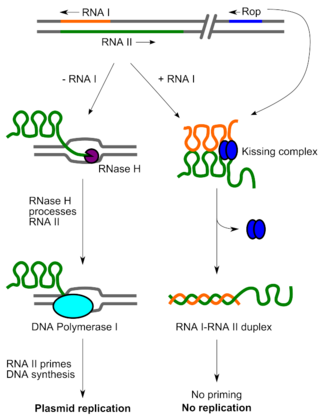
Rop is a small dimeric protein responsible for keeping the copy number of ColE1 family and related bacterial plasmids low in E. coli by increasing the speed of pairing between the preprimer RNA, RNA II, and its antisense RNA, RNA I. Structurally, Rop is a homodimeric four-helix bundle protein formed by the antiparallel interaction of two helix-turn-helix monomers. The Rop protein's structure has been solved to high resolution. Due to its small size and known structure, Rop has been used in protein design work to rearrange its helical topology and reengineer its loop regions. In general, the four-helix bundle has been extensively used in de novo protein design work as a simple model to understand the relationship between amino acid sequence and structure.

Emodepside is an anthelmintic drug that is effective against a number of gastrointestinal nematodes, is licensed for use in cats and belongs to the class of drugs known as the octadepsipeptides, a relatively new class of anthelmintic, which are suspected to achieve their anti-parasitic effect by a novel mechanism of action due to their ability to kill nematodes resistant to other anthelmintics.

CED-12 is a cytoplasmic, PH-domain containing adaptor protein found in Caenorhabditis elegans and Drosophila melanogaster. CED-12 is a homolog to the ELMO protein found in mammals. This protein is involved in Rac-GTPase activation, apoptotic cell phagocytosis, cell migration, and cytoskeletal rearrangements.

In molecular biology, the IMD domain is a BAR-like domain of approximately 250 amino acids found at the N-terminus in the insulin receptor tyrosine kinase substrate p53 (IRSp53/BAIAP2) and in the evolutionarily related IRSp53/MIM (MTSS1) family. In IRSp53, a ubiquitous regulator of the actin cytoskeleton, the IMD domain acts as conserved F-actin bundling domain involved in filopodium formation. Filopodium-inducing IMD activity is regulated by Cdc42 and Rac1 and is SH3-independent. The IRSp53/MIM family is a novel F-actin bundling protein family that includes invertebrate relatives:
Tc1/mariner is a class and superfamily of interspersed repeats DNA transposons. The elements of this class are found in all animals, including humans. They can also be found in protists and bacteria.

A single-pass membrane protein also known as single-spanning protein or bitopic protein is a transmembrane protein that spans the lipid bilayer only once. These proteins may constitute up to 50% of all transmembrane proteins, depending on the organism, and contribute significantly to the network of interactions between different proteins in cells, including interactions via transmembrane alpha helices. They usually include one or several water-soluble domains situated at the different sides of biological membranes, for example in single-pass transmembrane receptors. Some of them are small and serve as regulatory or structure-stabilizing subunits in large multi-protein transmembrane complexes, such as photosystems or the respiratory chain. A 2013 estimate identified about 1300 single-pass membrane proteins in the human genome.
















