Peter Michael Waterhouse is a British-Australian plant virologist and geneticist. He is a professor at the Queensland University of Technology and a Chief Investigator at the ARC Centre of Excellence for Plant Success in Nature and Agriculture.
Peter Michael Waterhouse is a British-Australian plant virologist and geneticist. He is a professor at the Queensland University of Technology and a Chief Investigator at the ARC Centre of Excellence for Plant Success in Nature and Agriculture.
Peter Waterhouse received his Bachelor of Science degree (1977) from the University of Newcastle upon Tyne, UK, and his Doctor of Philosophy degree (1981) from the Scottish Crop Research Institute at the University of Dundee. He joined CSIRO Plant Industry as a Research Scientist in 1981 [1] and was the leader of the virology program from 1989 to 1999 (excluding 1992). In 2008, he was awarded the Federation Fellowship and took up a professorship in the School of Molecular and Microbial Biosciences at the University of Sydney. Following the conclusion of this fellowship, Waterhouse left the University of Sydney and took up a professorial appointment at the Queensland University of Technology in 2014.
In the late 1990s, Waterhouse led the CSIRO group which discovered that gene silencing and transgene-mediated virus resistance in plants were induced and targeted by double-stranded RNAs.

Micro ribonucleic acid are small, single-stranded, non-coding RNA molecules containing 21–23 nucleotides. Found in plants, animals, and even some viruses, miRNAs are involved in RNA silencing and post-transcriptional regulation of gene expression. miRNAs base-pair to complementary sequences in messenger RNA (mRNA) molecules, then silence said mRNA molecules by one or more of the following processes:
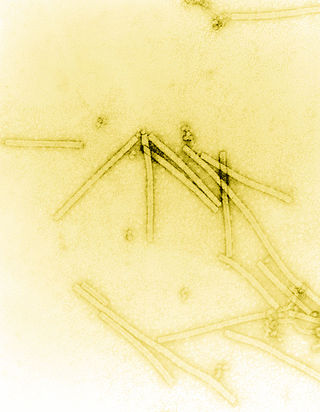
Tobacco mosaic virus (TMV) is a positive-sense single-stranded RNA virus species in the genus Tobamovirus that infects a wide range of plants, especially tobacco and other members of the family Solanaceae. The infection causes characteristic patterns, such as "mosaic"-like mottling and discoloration on the leaves. TMV was the first virus to be discovered. Although it was known from the late 19th century that a non-bacterial infectious disease was damaging tobacco crops, it was not until 1930 that the infectious agent was determined to be a virus. It is the first pathogen identified as a virus. The virus was crystallised by Wendell Meredith Stanley. It has a similar size to the largest synthetic molecule, known as PG5.
Gene silencing is the regulation of gene expression in a cell to prevent the expression of a certain gene. Gene silencing can occur during either transcription or translation and is often used in research. In particular, methods used to silence genes are being increasingly used to produce therapeutics to combat cancer and other diseases, such as infectious diseases and neurodegenerative disorders.

Dicer, also known as endoribonuclease Dicer or helicase with RNase motif, is an enzyme that in humans is encoded by the DICER1 gene. Being part of the RNase III family, Dicer cleaves double-stranded RNA (dsRNA) and pre-microRNA (pre-miRNA) into short double-stranded RNA fragments called small interfering RNA and microRNA, respectively. These fragments are approximately 20–25 base pairs long with a two-base overhang on the 3′-end. Dicer facilitates the activation of the RNA-induced silencing complex (RISC), which is essential for RNA interference. RISC has a catalytic component Argonaute, which is an endonuclease capable of degrading messenger RNA (mRNA).
The RNA-induced silencing complex, or RISC, is a multiprotein complex, specifically a ribonucleoprotein, which functions in gene silencing via a variety of pathways at the transcriptional and translational levels. Using single-stranded RNA (ssRNA) fragments, such as microRNA (miRNA), or double-stranded small interfering RNA (siRNA), the complex functions as a key tool in gene regulation. The single strand of RNA acts as a template for RISC to recognize complementary messenger RNA (mRNA) transcript. Once found, one of the proteins in RISC, Argonaute, activates and cleaves the mRNA. This process is called RNA interference (RNAi) and it is found in many eukaryotes; it is a key process in defense against viral infections, as it is triggered by the presence of double-stranded RNA (dsRNA).

Plant viruses are viruses that have the potential to affect plants. Like all other viruses, plant viruses are obligate intracellular parasites that do not have the molecular machinery to replicate without a host. Plant viruses can be pathogenic to vascular plants.
A transgene is a gene that has been transferred naturally, or by any of a number of genetic engineering techniques, from one organism to another. The introduction of a transgene, in a process known as transgenesis, has the potential to change the phenotype of an organism. Transgene describes a segment of DNA containing a gene sequence that has been isolated from one organism and is introduced into a different organism. This non-native segment of DNA may either retain the ability to produce RNA or protein in the transgenic organism or alter the normal function of the transgenic organism's genetic code. In general, the DNA is incorporated into the organism's germ line. For example, in higher vertebrates this can be accomplished by injecting the foreign DNA into the nucleus of a fertilized ovum. This technique is routinely used to introduce human disease genes or other genes of interest into strains of laboratory mice to study the function or pathology involved with that particular gene.
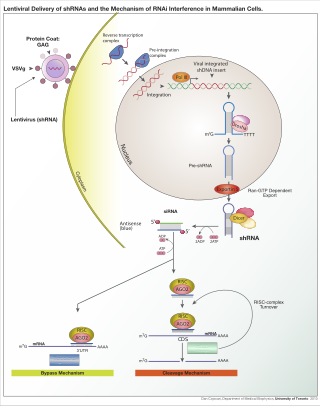
A short hairpin RNA or small hairpin RNA is an artificial RNA molecule with a tight hairpin turn that can be used to silence target gene expression via RNA interference (RNAi). Expression of shRNA in cells is typically accomplished by delivery of plasmids or through viral or bacterial vectors. shRNA is an advantageous mediator of RNAi in that it has a relatively low rate of degradation and turnover. However, it requires use of an expression vector, which has the potential to cause side effects in medicinal applications.
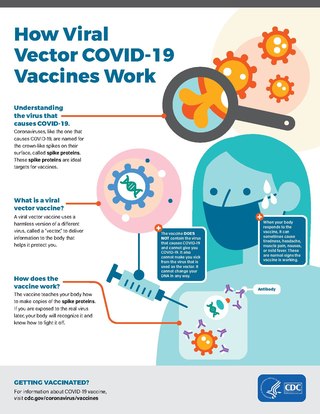
Viral vectors are modified viruses designed to deliver genetic material into cells. This process can be performed inside an organism or in cell culture. Viral vectors have widespread applications in basic research, agriculture, and medicine.

Gene delivery is the process of introducing foreign genetic material, such as DNA or RNA, into host cells. Gene delivery must reach the genome of the host cell to induce gene expression. Successful gene delivery requires the foreign gene delivery to remain stable within the host cell and can either integrate into the genome or replicate independently of it. This requires foreign DNA to be synthesized as part of a vector, which is designed to enter the desired host cell and deliver the transgene to that cell's genome. Vectors utilized as the method for gene delivery can be divided into two categories, recombinant viruses and synthetic vectors.

The Argonaute protein family, first discovered for its evolutionarily conserved stem cell function, plays a central role in RNA silencing processes as essential components of the RNA-induced silencing complex (RISC). RISC is responsible for the gene silencing phenomenon known as RNA interference (RNAi). Argonaute proteins bind different classes of small non-coding RNAs, including microRNAs (miRNAs), small interfering RNAs (siRNAs) and Piwi-interacting RNAs (piRNAs). Small RNAs guide Argonaute proteins to their specific targets through sequence complementarity, which then leads to mRNA cleavage, translation inhibition, and/or the initiation of mRNA decay.
RNA-induced transcriptional silencing (RITS) is a form of RNA interference by which short RNA molecules – such as small interfering RNA (siRNA) – trigger the downregulation of transcription of a particular gene or genomic region. This is usually accomplished by posttranslational modification of histone tails which target the genomic region for heterochromatin formation. The protein complex that binds to siRNAs and interacts with the methylated lysine 9 residue of histones H3 (H3K9me2) is the RITS complex.
RNA silencing or RNA interference refers to a family of gene silencing effects by which gene expression is negatively regulated by non-coding RNAs such as microRNAs. RNA silencing may also be defined as sequence-specific regulation of gene expression triggered by double-stranded RNA (dsRNA). RNA silencing mechanisms are conserved among most eukaryotes. The most common and well-studied example is RNA interference (RNAi), in which endogenously expressed microRNA (miRNA) or exogenously derived small interfering RNA (siRNA) induces the degradation of complementary messenger RNA. Other classes of small RNA have been identified, including piwi-interacting RNA (piRNA) and its subspecies repeat associated small interfering RNA (rasiRNA).

Sir David Charles Baulcombe is a British plant scientist and geneticist. As of October 2024 he was Head of Group, Gene Expression, in the Department of Plant Sciences at the University of Cambridge, and the Edward Penley Abraham Royal Society Research Professor and Regius Professor of Botany Emeritus at Cambridge. He held the Regius botany chair in that department from 2007 to 2020.

miR-122 is a miRNA that is conserved among vertebrate species. miR-122 is not present in invertebrates, and no close paralogs of miR-122 have been detected. miR-122 is highly expressed in the liver, where it has been implicated as a regulator of fatty-acid metabolism in mouse studies. Reduced miR-122 levels are associated with hepatocellular carcinoma. miR-122 also plays an important positive role in the regulation of hepatitis C virus replication.

RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by other names, including co-suppression, post-transcriptional gene silencing (PTGS), and quelling. The detailed study of each of these seemingly different processes elucidated that the identity of these phenomena were all actually RNAi. Andrew Fire and Craig C. Mello shared the 2006 Nobel Prize in Physiology or Medicine for their work on RNAi in the nematode worm Caenorhabditis elegans, which they published in 1998. Since the discovery of RNAi and its regulatory potentials, it has become evident that RNAi has immense potential in suppression of desired genes. RNAi is now known as precise, efficient, stable and better than antisense therapy for gene suppression. Antisense RNA produced intracellularly by an expression vector may be developed and find utility as novel therapeutic agents.
Adenovirus varieties have been explored extensively as a viral vector for gene therapy and also as an oncolytic virus.

RNA-directed DNA methylation (RdDM) is a biological process in which non-coding RNA molecules direct the addition of DNA methylation to specific DNA sequences. The RdDM pathway is unique to plants, although other mechanisms of RNA-directed chromatin modification have also been described in fungi and animals. To date, the RdDM pathway is best characterized within angiosperms, and particularly within the model plant Arabidopsis thaliana. However, conserved RdDM pathway components and associated small RNAs (sRNAs) have also been found in other groups of plants, such as gymnosperms and ferns. The RdDM pathway closely resembles other sRNA pathways, particularly the highly conserved RNAi pathway found in fungi, plants, and animals. Both the RdDM and RNAi pathways produce sRNAs and involve conserved Argonaute, Dicer and RNA-dependent RNA polymerase proteins.
Emily Bernstein is a professor at Mount Sinai School of Medicine known for her research on RNA interference, epigenetics, and cancer, especially melanoma.
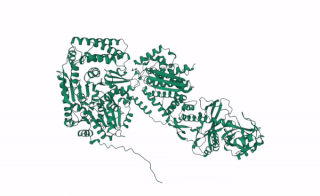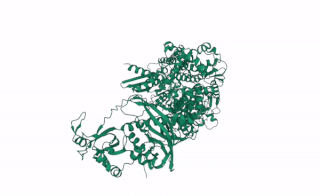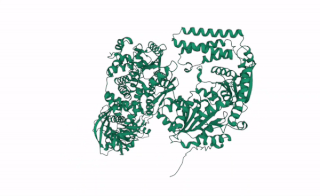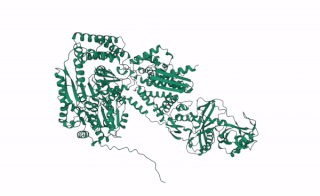
DCL2 is a gene in plants that codes for the DCL2 protein, a ribonuclease III enzyme involved in processing exogenous double-stranded RNA (dsRNA) into 22 nucleotide small interference RNAs (siRNAs).