External links
- Philippe Horvath on ORCID
- Philippe Horvath on Researcher ID
- Philippe Horvath on the Copains d’avant social network
Philippe Horvath | |
|---|---|
| Born | April 17, 1970 |
| Nationality | French |
| Education | Université Louis Pasteur, Strasbourg |
| Occupation | Scientist |
| Employer(s) | DuPont Nutrition and Health |
Philippe Horvath is a French scientist working for DuPont Nutrition and Health. His work was integral to the development of CRISPR-Cas, a versatile biochemical method for targeted genetic engineering. For this work, he was awarded the 2015 Massry Prize along with Emmanuelle Charpentier and Jennifer Doudna, as well as the 2016 Canada Gairdner International Award, with his Massry co-laureates in addition to Feng Zhang, Rodolphe Barrangou, Anthony Fauci, and Frank Plummer.
After attending school in Colmar, Horvath studied cellular and molecular biology at the Université Louis Pasteur, Strasbourg, where he obtained a Master's in 1996 and a Ph.D. in 2000. After graduation, he went to the Department of Research and Development of Rhodia Food (formerly Rhône-Poulenc) in Dangé-Saint-Romain, where he worked to develop molecular biology techniques for bacterial strain screening, microbial identification, and typing of lactic acid bacteria and their phages. [1] In 2004, Rhodia Food was acquired by Danisco, and Philippe was promoted to senior scientist in 2006. The division was later purchased by DuPont in 2011, and Horvath was appointed an Associate to the DuPort Fellows Forum in 2014, and a DuPont Nutrition & Health Technical Fellow in 2015. [1]
Since late 2002, Philippe's research activities have centered around CRISPR (clustered regularly interspaced short palindromic repeats). Early work was stimulated by with the aim of improving the durability of bacterial starter cultures to improve the manufacture of cheese and ice cream, [2] particularly efforts to address bacteriophages---viruses that infect bacteria. Horvath explored sections in the bacterial genome with clustered regularly interspaced short palindromic repeats, both for their utility in differentiating between strains, and because of their role in the bacterial immune system. As this prokaryotic viral defense mechanism was understood, it was recognized that CRISPRs could be used with specific endonuclease enzymes for genome editing and gene regulation. As of 2016, Philippe has been the co-inventor of 95 patents and/or applications, and co-author of 31 peer-reviewed research articles. [1]
A restriction enzyme, restriction endonuclease, REase, ENase orrestrictase is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each sugar-phosphate backbone of the DNA double helix.
Gene knockouts are a widely used genetic engineering technique that involves the targeted removal or inactivation of a specific gene within an organism's genome. This can be done through a variety of methods, including homologous recombination, CRISPR-Cas9, and TALENs.
Gene knockdown is an experimental technique by which the expression of one or more of an organism's genes is reduced. The reduction can occur either through genetic modification or by treatment with a reagent such as a short DNA or RNA oligonucleotide that has a sequence complementary to either gene or an mRNA transcript.

CRISPR is a family of DNA sequences found in the genomes of prokaryotic organisms such as bacteria and archaea. These sequences are derived from DNA fragments of bacteriophages that had previously infected the prokaryote. They are used to detect and destroy DNA from similar bacteriophages during subsequent infections. Hence these sequences play a key role in the antiviral defense system of prokaryotes and provide a form of acquired immunity. CRISPR is found in approximately 50% of sequenced bacterial genomes and nearly 90% of sequenced archaea.
A guide RNA (gRNA) is a piece of RNA that functions as a guide for RNA- or DNA-targeting enzymes, with which it forms complexes. Very often these enzymes will delete, insert or otherwise alter the targeted RNA or DNA. They occur naturally, serving important functions, but can also be designed to be used for targeted editing, such as with CRISPR-Cas9 and CRISPR-Cas12.

Stenotrophomonas is a genus of Gram-negative bacteria, comprising at least ten species. The main reservoirs of Stenotrophomonas are soil and plants. Stenotrophomonas species range from common soil organisms to opportunistic human pathogens ; the molecular taxonomy of the genus is still somewhat unclear.

In Molecular biology, an insert is a piece of DNA that is inserted into a larger DNA vector by a recombinant DNA technique, such as ligation or recombination. This allows it to be multiplied, selected, further manipulated or expressed in a host organism.

Genome editing, or genome engineering, or gene editing, is a type of genetic engineering in which DNA is inserted, deleted, modified or replaced in the genome of a living organism. Unlike early genetic engineering techniques that randomly inserts genetic material into a host genome, genome editing targets the insertions to site-specific locations. The basic mechanism involved in genetic manipulations through programmable nucleases is the recognition of target genomic loci and binding of effector DNA-binding domain (DBD), double-strand breaks (DSBs) in target DNA by the restriction endonucleases, and the repair of DSBs through homology-directed recombination (HDR) or non-homologous end joining (NHEJ).
In molecular biology, trans-activating crispr RNA (tracrRNA) is a small trans-encoded RNA. It was first discovered by Emmanuelle Charpentier in her study of human pathogen Streptococcus pyogenes, a type of bacteria that causes harm to humanity. In bacteria and archaea; CRISPR-Cas constitute an RNA-mediated defense system which protects against viruses and plasmids. This defensive pathway has three steps. First a copy of the invading nucleic acid is integrated into the CRISPR locus. Next, CRISPR RNAs (crRNAs) are transcribed from this CRISPR locus. The crRNAs are then incorporated into effector complexes, where the crRNA guides the complex to the invading nucleic acid and the Cas proteins degrade this nucleic acid. There are several CRISPR system subtypes.

Genetic engineering techniques allow the modification of animal and plant genomes. Techniques have been devised to insert, delete, and modify DNA at multiple levels, ranging from a specific base pair in a specific gene to entire genes. There are a number of steps that are followed before a genetically modified organism (GMO) is created. Genetic engineers must first choose what gene they wish to insert, modify, or delete. The gene must then be isolated and incorporated, along with other genetic elements, into a suitable vector. This vector is then used to insert the gene into the host genome, creating a transgenic or edited organism.

Cas9 is a 160 kilodalton protein which plays a vital role in the immunological defense of certain bacteria against DNA viruses and plasmids, and is heavily utilized in genetic engineering applications. Its main function is to cut DNA and thereby alter a cell's genome. The CRISPR-Cas9 genome editing technique was a significant contributor to the Nobel Prize in Chemistry in 2020 being awarded to Emmanuelle Charpentier and Jennifer Doudna.

CRISPR interference (CRISPRi) is a genetic perturbation technique that allows for sequence-specific repression of gene expression in prokaryotic and eukaryotic cells. It was first developed by Stanley Qi and colleagues in the laboratories of Wendell Lim, Adam Arkin, Jonathan Weissman, and Jennifer Doudna. Sequence-specific activation of gene expression refers to CRISPR activation (CRISPRa).

CRISPR-associated protein 1 (cas1) is one of the two universally conserved proteins found in the CRISPR prokaryotic immune defense system. Cas1 is a metal-dependent DNA-specific endonuclease that produces double-stranded DNA fragments. Cas1 forms a stable complex with the other universally conserved CRISPR-associated protein, cas2, which is essential to spacer acquisition for CRISPR systems.
Yoshizumi Ishino is a Japanese molecular biologist, known for his discovering the DNA sequence of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR).

Cas12a is a subtype of Cas12 proteins and an RNA-guided endonuclease that forms part of the CRISPR system in some bacteria and archaea. It originates as part of a bacterial immune mechanism, where it serves to destroy the genetic material of viruses and thus protect the cell and colony from viral infection. Cas12a and other CRISPR associated endonucleases use an RNA to target nucleic acid in a specific and programmable matter. In the organisms from which it originates, this guide RNA is a copy of a piece of foreign nucleic acid that previously infected the cell.
Rodolphe Barrangou is the Todd R. Klaenhammer Distinguished Professor in Probiotics Research in the Department of Food, Bioprocessing and Nutrition Sciences at North Carolina State University; Co-Founder and Chief Executive Officer of CRISPR Biotechnologies; Co-Founder and Chief Scientific Officer of Ancilia Biosciences; Co-Founder, President and Chief Scientific Officer of TreeCo; and Co-Founder and member of the Scientific Advisory Board of Intellia Therapeutics. His research focuses on CRISPR-Cas9 in bacteria.

Francisco Juan Martínez Mojica is a Spanish molecular biologist and microbiologist at the University of Alicante in Spain. He is known for his discovery of repetitive, functional DNA sequences in bacteria which he named CRISPR. These were later developed into the first widespread genome editing tool, CRISPR-Cas9.
Samira Kiani is an Associate Professor in the department of Pathology of University of Pittsburgh School of Medicine and Pittsburgh Liver Research Center. Formerly, she was a Health Systems Engineer at Arizona State University. Her work combines Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR) with synthetic biology. She is a 2019 AAAS Leshner Fellow.
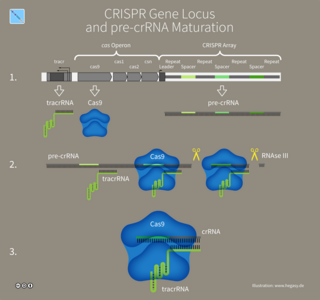
CRISPR RNA or crRNA is a RNA transcript from the CRISPR locus. CRISPR-Cas is an adaptive immune system found in bacteria and archaea to protect against mobile genetic elements, like viruses, plasmids, and transposons. The CRISPR locus contains a series of repeats interspaced with unique spacers. These unique spacers can be acquired from MGEs.