
A pilus is a hair-like appendage found on the surface of many bacteria and archaea. The terms pilus and fimbria can be used interchangeably, although some researchers reserve the term pilus for the appendage required for bacterial conjugation. All conjugative pili are primarily composed of pilin – fibrous proteins, which are oligomeric.

Borrelia burgdorferi is a bacterial species of the spirochete class in the genus Borrelia, and is one of the causative agents of Lyme disease in humans. Along with a few similar genospecies, some of which also cause Lyme disease, it makes up the species complex of Borrelia burgdorferi sensu lato. The complex currently comprises 20 accepted and 3 proposed genospecies. B. burgdorferi sensu stricto exists in North America and Eurasia and until 2016 was the only known cause of Lyme disease in North America. Borrelia species are Gram-negative.

Pseudomonas aeruginosa is a common encapsulated, Gram-negative, aerobic–facultatively anaerobic, rod-shaped bacterium that can cause disease in plants and animals, including humans. A species of considerable medical importance, P. aeruginosa is a multidrug resistant pathogen recognized for its ubiquity, its intrinsically advanced antibiotic resistance mechanisms, and its association with serious illnesses – hospital-acquired infections such as ventilator-associated pneumonia and various sepsis syndromes. P. aeruginosa is able to selectively inhibit various antibiotics from penetrating its outer membrane - and has high resistance to several antibiotics, according to the World Health Organization P. aeruginosa poses one of the greatest threats to humans in terms of antibiotic resistance.
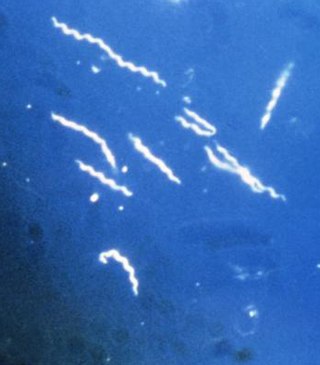
Lyme disease, or borreliosis, is caused by spirochetal bacteria from the genus Borrelia, which has 52 known species. Three main species are the main causative agents of the disease in humans, while a number of others have been implicated as possibly pathogenic. Borrelia species in the species complex known to cause Lyme disease are collectively called Borrelia burgdorferisensu lato (s.l.) not to be confused with the single species in that complex Borrelia burgdorferi sensu stricto which is responsible for nearly all cases of Lyme disease in North America.

Swarming motility is a rapid and coordinated translocation of a bacterial population across solid or semi-solid surfaces, and is an example of bacterial multicellularity and swarm behaviour. Swarming motility was first reported by Jorgen Henrichsen and has been mostly studied in genus Serratia, Salmonella, Aeromonas, Bacillus, Yersinia, Pseudomonas, Proteus, Vibrio and Escherichia.

Virulence-related outer membrane proteins, or outer surface proteins (Osp) in some contexts, are expressed in the outer membrane of gram-negative bacteria and are essential to bacterial survival within macrophages and for eukaryotic cell invasion.

Bacterial motility is the ability of bacteria to move independently using metabolic energy. Most motility mechanisms that evolved among bacteria also evolved in parallel among the archaea. Most rod-shaped bacteria can move using their own power, which allows colonization of new environments and discovery of new resources for survival. Bacterial movement depends not only on the characteristics of the medium, but also on the use of different appendages to propel. Swarming and swimming movements are both powered by rotating flagella. Whereas swarming is a multicellular 2D movement over a surface and requires the presence of surfactants, swimming is movement of individual cells in liquid environments.

Cyclic di-GMP is a second messenger used in signal transduction in a wide variety of bacteria. Cyclic di-GMP is not known to be used by archaea, and has only been observed in eukaryotes in Dictyostelium. The biological role of cyclic di-GMP was first uncovered when it was identified as an allosteric activator of a cellulose synthase found in Gluconacetobacter xylinus in order to produce microbial cellulose.

Cyclic di-GMP-I riboswitches are a class of riboswitch that specifically bind cyclic di-GMP, which is a second messenger that is used in a variety of microbial processes including virulence, motility and biofilm formation. Cyclic di-GMP-I riboswitches were originally identified by bioinformatics as a conserved RNA-like structure called the "GEMM motif". These riboswitches are present in a wide variety of bacteria, and are most common in Clostridia and certain varieties of Pseudomonadota. The riboswitches are present in pathogens such as Clostridium difficile, Vibrio cholerae and Bacillus anthracis. Geobacter uraniumreducens is predicted to have 30 instances of this riboswitch in its genome. A bacteriophage that infects C. difficile is predicted to carry a cyclic di-GMP-I riboswitch, which it might use to detect and exploit the physiological state of bacteria that it infects.
Motility protein A, MotA, is a bacterial protein that is encoded by the motA gene. It is a component of the flagellar motor. More specifically, MotA and MotB make the stator of a H+ driven bacterial flagella and surround the rotor as a ring of about 8–10 particles. MotA and MotB are integral membrane proteins. MotA has four transmembrane domains.

The fimbrial usher protein is involved in biogenesis of the pilus in Gram-negative bacteria. The biogenesis of some fimbriae requires a two-component assembly and transport system which is composed of a periplasmic chaperone and a pore-forming outer membrane protein which has been termed a molecular 'usher'; this is the chaperone-usher pathway.

CobB is a bacterial protein that belongs to the sirtuin family, a broadly conserved family of NAD+-dependent protein deacetylases.
In enzymology, diguanylate cyclase, also known as diguanylate kinase, is an enzyme that catalyzes the chemical reaction:
In molecular biology, the flagellar motor switch protein(Flig) is one of three proteins in certain bacteria coded for by the gene fliG. The other two proteins are FliN coded for by fliN, and FliM coded for by fliM. The protein complex regulates the direction of flagellar rotation and hence controls swimming behaviour. The switch is a complex apparatus that responds to signals transduced by the chemotaxis sensory signalling system during chemotactic behaviour. CheY, the chemotaxis response regulator, is believed to act directly on the switch to induce a switch in the flagellar motor direction of rotation.
In molecular biology, the GGDEF domain is a protein domain which appears to be ubiquitous in bacteria and is often linked to a regulatory domain, such as a phosphorylation receiver or oxygen sensing domain. Its function is to act as a diguanylate cyclase and synthesize cyclic di-GMP, which is used as an intracellular signalling molecule in a wide variety of bacteria. Enzymatic activity can be strongly influenced by the adjacent domains. Processes regulated by this domain include exopolysaccharide synthesis, biofilm formation, motility and cell differentiation.

A response regulator is a protein that mediates a cell's response to changes in its environment as part of a two-component regulatory system. Response regulators are coupled to specific histidine kinases which serve as sensors of environmental changes. Response regulators and histidine kinases are two of the most common gene families in bacteria, where two-component signaling systems are very common; they also appear much more rarely in the genomes of some archaea, yeasts, filamentous fungi, and plants. Two-component systems are not found in metazoans.
The archaellum is a unique structure on the cell surface of many archaea that allows for swimming motility. The archaellum consists of a rigid helical filament that is attached to the cell membrane by a molecular motor. This molecular motor – composed of cytosolic, membrane, and pseudo-periplasmic proteins – is responsible for the assembly of the filament and, once assembled, for its rotation. The rotation of the filament propels archaeal cells in liquid medium, in a manner similar to the propeller of a boat. The bacterial analog of the archaellum is the flagellum, which is also responsible for their swimming motility and can also be compared to a rotating corkscrew. Although the movement of archaella and flagella is sometimes described as "whip-like", this is incorrect, as only cilia from Eukaryotes move in this manner. Indeed, even "flagellum" is a misnomer, as bacterial flagella also work as propeller-like structures.
Chaperone-usher fimbriae (CU) are linear, unbranching, outer-membrane pili secreted by gram-negative bacteria through the chaperone-usher system rather than through type IV secretion or extracellular nucleation systems. These fimbriae are built up out of modular pilus subunits, which are transported into the periplasm in a Sec dependent manner. Chaperone-usher secreted fimbriae are important pathogenicity factors facilitating host colonisation, localisation and biofilm formation in clinically important species such as uropathogenic Escherichia coli and Pseudomonas aeruginosa.

Twitching motility is a form of crawling bacterial motility used to move over surfaces. Twitching is mediated by the activity of hair-like filaments called type IV pili which extend from the cell's exterior, bind to surrounding solid substrates, and retract, pulling the cell forwards in a manner similar to the action of a grappling hook. The name twitching motility is derived from the characteristic jerky and irregular motions of individual cells when viewed under the microscope. It has been observed in many bacterial species, but is most well studied in Pseudomonas aeruginosa, Neisseria gonorrhoeae and Myxococcus xanthus. Active movement mediated by the twitching system has been shown to be an important component of the pathogenic mechanisms of several species.
CsgD is a transcription and response regulator protein referenced to as the master modulator of bacterial biofilm development. In E. coli cells, CsgD is tasked with aiding the transition from planktonic cell motility to the stationary phase of biofilm formation, in response to environmental growth factors. A transcription analysis assay illustrated a heightened decrease in CsgD's DNA-binding capacity when phosphorylated at A.A. D59 of the protein's primary sequence. Therefore, in the protein's active form (unphosphorylated), CsgD is capable of carrying out its normal functions of regulating curli proteins (fimbria) and producing ECM polysaccharides (cellulose). Following a promoter-lacZ fusion assay of CsgD binding to specific target sites on E. coli's genome, two classes of binding targets were identified: group I genes and group II genes. The group I genes, akin to fliE and yhbT, exhibit repressed transcription following their interaction with CsgD, whilst group II genes, including yccT and adrA, illustrated active functionality. Other group I operons that illustrate repressed transcription include fliE and fliEFGH, for motile flagellum formation. Other group II genes, imperative to the transition towards stationary biofilm development, include csgBA, encoding for curli fimbriae, and adrA, encoding for the synthesis of cyclic diguanylate. In this context, c-di-GMP functions as a bacterial secondary messenger, enhancing the production of extracellular cellulose and impeding flagellum production and rotation.















