
Riboflavin, also known as vitamin B2, is a vitamin found in food and sold as a dietary supplement. It is essential to the formation of two major coenzymes, flavin mononucleotide and flavin adenine dinucleotide. These coenzymes are involved in energy metabolism, cellular respiration, and antibody production, as well as normal growth and development. The coenzymes are also required for the metabolism of niacin, vitamin B6, and folate. Riboflavin is prescribed to treat corneal thinning, and taken orally, may reduce the incidence of migraine headaches in adults.

In biochemistry, a kinase is an enzyme that catalyzes the transfer of phosphate groups from high-energy, phosphate-donating molecules to specific substrates. This process is known as phosphorylation, where the high-energy ATP molecule donates a phosphate group to the substrate molecule. This transesterification produces a phosphorylated substrate and ADP. Conversely, it is referred to as dephosphorylation when the phosphorylated substrate donates a phosphate group and ADP gains a phosphate group. These two processes, phosphorylation and dephosphorylation, occur four times during glycolysis.

Flavins refers generally to the class of organic compounds containing the tricyclic heterocycle isoalloxazine or its isomer alloxazine, and derivatives thereof. The biochemical source of flavin is the vitamin riboflavin. The flavin moiety is often attached with an adenosine diphosphate to form flavin adenine dinucleotide (FAD), and, in other circumstances, is found as flavin mononucleotide, a phosphorylated form of riboflavin. It is in one or the other of these forms that flavin is present as a prosthetic group in flavoproteins.
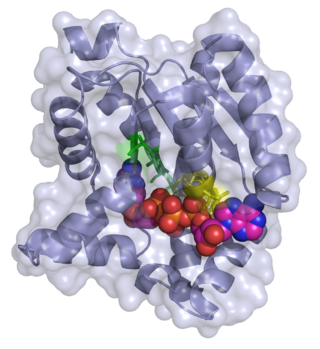
Adenylate kinase is a phosphotransferase enzyme that catalyzes the interconversion of the various adenosine phosphates. By constantly monitoring phosphate nucleotide levels inside the cell, ADK plays an important role in cellular energy homeostasis.

In biochemistry, flavin adenine dinucleotide (FAD) is a redox-active coenzyme associated with various proteins, which is involved with several enzymatic reactions in metabolism. A flavoprotein is a protein that contains a flavin group, which may be in the form of FAD or flavin mononucleotide (FMN). Many flavoproteins are known: components of the succinate dehydrogenase complex, α-ketoglutarate dehydrogenase, and a component of the pyruvate dehydrogenase complex.

Flavin mononucleotide (FMN), or riboflavin-5′-phosphate, is a biomolecule produced from riboflavin (vitamin B2) by the enzyme riboflavin kinase and functions as the prosthetic group of various oxidoreductases, including NADH dehydrogenase, as well as cofactor in biological blue-light photo receptors. During the catalytic cycle, a reversible interconversion of the oxidized (FMN), semiquinone (FMNH•), and reduced (FMNH2) forms occurs in the various oxidoreductases. FMN is a stronger oxidizing agent than NAD and is particularly useful because it can take part in both one- and two-electron transfers. In its role as blue-light photo receptor, (oxidized) FMN stands out from the 'conventional' photo receptors as the signaling state and not an E/Z isomerization.
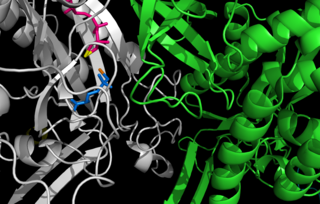
Glutamine synthetase (GS) is an enzyme that plays an essential role in the metabolism of nitrogen by catalyzing the condensation of glutamate and ammonia to form glutamine:

Flavoproteins are proteins that contain a nucleic acid derivative of riboflavin. These proteins are involved in a wide array of biological processes, including removal of radicals contributing to oxidative stress, photosynthesis, and DNA repair. The flavoproteins are some of the most-studied families of enzymes.

Amino acid synthesis is the set of biochemical processes by which the amino acids are produced. The substrates for these processes are various compounds in the organism's diet or growth media. Not all organisms are able to synthesize all amino acids. For example, humans can synthesize 11 of the 20 standard amino acids. These 11 are called the non-essential amino acids).
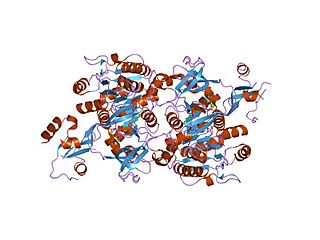
The long chain fatty acyl-CoA ligase is an enzyme of the ligase family that activates the oxidation of complex fatty acids. Long chain fatty acyl-CoA synthetase catalyzes the formation of fatty acyl-CoA by a two-step process proceeding through an adenylated intermediate. The enzyme catalyzes the following reaction,

Adenylylation, more commonly known as AMPylation, is a process in which an adenosine monophosphate (AMP) molecule is covalently attached to the amino acid side chain of a protein. This covalent addition of AMP to a hydroxyl side chain of the protein is a post-translational modification. Adenylylation involves a phosphodiester bond between a hydroxyl group of the molecule undergoing adenylylation, and the phosphate group of the adenosine monophosphate nucleotide. Enzymes that are capable of catalyzing this process are called AMPylators.
In enzymology, a FAD diphosphatase (EC 3.6.1.18) is an enzyme that catalyzes the chemical reaction
In enzymology, a FMN adenylyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a nucleoside-phosphate kinase is an enzyme that catalyzes the chemical reaction
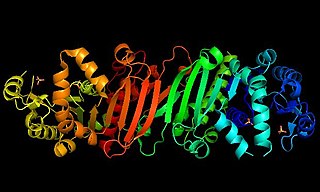
Phosphoribulokinase (PRK) (EC 2.7.1.19) is an essential photosynthetic enzyme that catalyzes the ATP-dependent phosphorylation of ribulose 5-phosphate (RuP) into ribulose 1,5-bisphosphate (RuBP), both intermediates in the Calvin Cycle. Its main function is to regenerate RuBP, which is the initial substrate and CO2-acceptor molecule of the Calvin Cycle. PRK belongs to the family of transferase enzymes, specifically those transferring phosphorus-containing groups (phosphotransferases) to an alcohol group acceptor. Along with ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCo), phosphoribulokinase is unique to the Calvin Cycle. Therefore, PRK activity often determines the metabolic rate in organisms for which carbon fixation is key to survival. Much initial work on PRK was done with spinach leaf extracts in the 1950s; subsequent studies of PRK in other photosynthetic prokaryotic and eukaryotic organisms have followed. The possibility that PRK might exist was first recognized by Weissbach et al. in 1954; for example, the group noted that carbon dioxide fixation in crude spinach extracts was enhanced by the addition of ATP. The first purification of PRK was conducted by Hurwitz and colleagues in 1956.
ATP + Mg2+ - D-ribulose 5-phosphate ADP + D-ribulose 1,5-bisphosphate

In enzymology, a riboflavin kinase is an enzyme that catalyzes the chemical reaction
In enzymology, a streptomycin 3"-adenylyltransferase is an enzyme that catalyzes the chemical reaction:

In enzymology, a sulfate adenylyltransferase is an enzyme that catalyzes the chemical reaction

Triokinase/FMN cyclase is an enzyme that in humans is encoded by the DAK gene.















