
Pseudomonas is a genus of Gram-negative, Gammaproteobacteria, belonging to the family Pseudomonadaceae and containing 191 described species. The members of the genus demonstrate a great deal of metabolic diversity and consequently are able to colonize a wide range of niches. Their ease of culture in vitro and availability of an increasing number of Pseudomonas strain genome sequences has made the genus an excellent focus for scientific research; the best studied species include P. aeruginosa in its role as an opportunistic human pathogen, the plant pathogen P. syringae, the soil bacterium P. putida, and the plant growth-promoting P. fluorescens, P. lini, P. migulae, and P. graminis.

A plant canker is a small area of dead tissue, which grows slowly, often over years. Some cankers are of only minor consequence, but others are ultimately lethal and therefore can have major economic implications for agriculture and horticulture. Their causes include a wide range of organisms as fungi, bacteria, mycoplasmas and viruses. The majority of canker-causing organisms are bound to a unique host species or genus, but a few will attack other plants. Weather and animals can spread canker, thereby endangering areas that have only slight amount of canker.
Phytotoxins are substances that are poisonous or toxic to the growth of plants. Phytotoxic substances may result from human activity, as with herbicides, or they may be produced by plants, by microorganisms, or by naturally occurring chemical reactions.
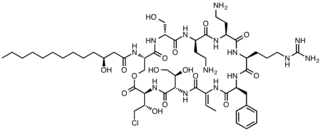
Syringomycin E is a member of a class of lipodepsinonapeptide molecules that are secreted by the plant pathogen Pseudomonas syringae. Lipodepsinonapeptides comprise a closed ring of nine nonribosomally synthesized amino acids bonded to a fatty acid hydrocarbon tail. A commonly encountered pathovar (pv) of P. syringae is P. syringae pv syringae, which secretes a number of closely related forms of the molecule. Syringomycins are virulence determinants, which means that their secretion is required for the manifestation of disease symptoms on a number of stone fruit crop plants.
The gene-for-gene relationship was discovered by Harold Henry Flor who was working with rust of flax. Flor showed that the inheritance of both resistance in the host and parasite ability to cause disease is controlled by pairs of matching genes. One is a plant gene called the resistance (R) gene. The other is a parasite gene called the avirulence (Avr) gene. Plants producing a specific R gene product are resistant towards a pathogen that produces the corresponding Avr gene product. Gene-for-gene relationships are a widespread and very important aspect of plant disease resistance. Another example can be seen with Lactuca serriola versus Bremia lactucae.

Pseudomonas syringae is a rod-shaped, Gram-negative bacterium with polar flagella. As a plant pathogen, it can infect a wide range of species, and exists as over 50 different pathovars, all of which are available to researchers from international culture collections such as the NCPPB, ICMP, and others.
Pseudomonas avellanae is a Gram-negative plant pathogenic bacterium. It is the causal agent of bacterial canker of hazelnut. Based on 16S rRNA analysis, P. avellanae has been placed in the P. syringae group. This species was once included as a pathovar of Pseudomonas syringae, but following DNA-DNA hybridization, it was instated as a separate species. Following ribotypical analysis Pseudomonas syringae pv. theae was incorporated into this species.

Pseudomonas savastanoi is a gram-negative plant pathogenic bacterium that infects a variety of plants. It was once considered a pathovar of Pseudomonas syringae, but following DNA-relatedness studies, it was instated as a new species. It is named after Savastano, a worker who proved between 1887 and 1898 that olive knot are caused by bacteria.
Pseudomonas viridiflava is a fluorescent, Gram-negative, soil bacterium that is pathogenic to plants. It was originally isolated from the dwarf or runner bean, in Switzerland. Based on 16S rRNA analysis, P. viridiflava has been placed in the P. syringae group. Following ribotypical analysis misidentified strains of Pseudomonas syringae pv. ribicola and Pseudomonas syringae pv. primulae were incorporated into this species. This pathogen causes bacterial blight of Kiwifruit.
Pseudomonas amygdali is a Gram-negative plant pathogenic bacterium. It is named after its ability to cause disease on almond trees. Different analyses, including 16S rRNA analysis, DNA-DNA hybridization, and MLST clearly placed P. amygdali in the P. syringae group together with the species Pseudomonas ficuserectae and Pseudomonas meliae, and 27 pathovars of Pseudomonas syringae/Pseudomonas savastanoi, constituting a single, well-defined phylogenetic group which should be considered as a single species. This phylogenetic group has not been formally named because of the lack of reliable means to differentiate it phenotypically from closely related species, and it is currently known as either genomospecies 2 or phylogroup 3. When it is formally named, the correct name for this new species should be Pseudomonas amygdali, which takes precedence over all the other names of taxa from this group, including Pseudomonas savastanoi, which is and inadequate and confusing name whose use is not recommended.

Pseudomonas cannabina is a gray, Gram-negative, fluorescent, motile, flagellated, aerobic bacterium that causes leaf and stem rot of hemp, from which it derives its name. It was formerly classified as a pathovar of Pseudomonas syringae, but following ribotypical analysis, it was reinstated as a species. The type strain is CFBP 2341.
Pseudomonas tremae is a white, Gram-negative, non-fluorescent, motile, flagellated, aerobic bacterium that infects Trema orientalis, from which it derives its name. It was formerly classified as a pathovar of Pseudomonas syringae, but following ribotypical analysis, it was instated as a species. The type strain is CFBP 3229.
Pseudomonas coronafaciens is a Gram-negative bacterium that is pathogenic to several plant species. Following ribotypical analysis several pathovars of Pseudomonas syringae were incorporated into this species.

Xanthomonas is a genus of bacteria, many of which cause plant diseases. There are at least 27 plant associated Xanthomonas spp., that all together infect at least 400 plant species. Different species typically have specific host and/or tissue range and colonization strategies.
"Pseudomonas helianthi" is a Gram-negative plant pathogenic bacterium that infects a variety of plants. It was once considered a pathovar of Pseudomonas syringae, but following DNA-relatedness studies, it was recognized as a separate species and P. syringae pv. tagetis was incorporated into it, as well. Since no official name has yet been given, it is referred to by the epithet 'Pseudomonas helianthi' .

Prunus necrotic ringspot virus (PNRSV) is a plant pathogenic virus causing ring spot diseases affecting species of the genus Prunus, as well as other species such as rose and hops. PNRSV is found worldwide due to easy transmission through plant propagation methods and infected seed. The virus is in the family Bromoviridae and genus Ilarvirus. Synonyms of PNRSV include European plum line pattern virus, hop B virus, hop C virus, plum line pattern virus, sour cherry necrotic ringspot virus, and peach ringspot virus.

The gyrA RNA motif is a conserved RNA structure identified by bioinformatics. The RNAs are present in multiple species of bacteria within the order Pseudomonadales. This order contains the genus Pseudomonas, which includes the opportunistic human pathogen Pseudomonas aeruginosa and Pseudomonas syringae, a plant pathogen.
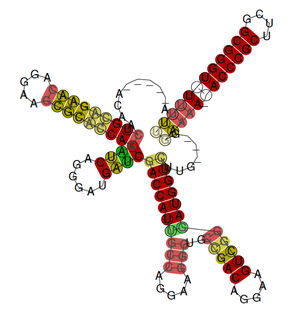
The rsmX gene is part of the Rsm/Csr family of non-coding RNAs (ncRNAs). Members of the Rsm/Csr family are present in a diverse range of bacteria, including Escherichia coli, Erwinia, Salmonella, Vibrio and Pseudomonas. These ncRNAs act by sequestering translational repressor proteins, called RsmA, activating expression of downstream genes that would normally be blocked by the repressors. Sequestering of target proteins is dependent upon exposed GGA motifs in the stem loops of the ncRNAs. Typically, the activated genes are involved in secondary metabolism, biofilm formation and motility.
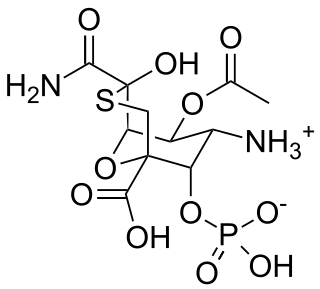
Tagetitoxin (TGT) is a bacterial phytotoxin produced by Pseudomonas syringae pv. tagetis.
Pseudomonas protegens are widespread Gram-negative, plant-protecting bacteria. Some of the strains of this novel bacterial species previously belonged to P. fluorescens. They were reclassified since they seem to cluster separately from other fluorescent Pseudomonas species. P. protegens is phylogenetically related to the Pseudomonas species complexes P. fluorescens, P. chlororaphis, and P. syringae. The bacterial species characteristically produces the antimicrobial compounds pyoluteorin and 2,4-diacetylphloroglucinol (DAPG) which are active against various plant pathogens.











