Related Research Articles

Helicases are a class of enzymes thought to be vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separating two hybridized nucleic acid strands, using energy from ATP hydrolysis. There are many helicases, representing the great variety of processes in which strand separation must be catalyzed. Approximately 1% of eukaryotic genes code for helicases.

Exodeoxyribonuclease V is an enzyme of E. coli that initiates recombinational repair from potentially lethal double strand breaks in DNA which may result from ionizing radiation, replication errors, endonucleases, oxidative damage, and a host of other factors. The RecBCD enzyme is both a helicase that unwinds, or separates the strands of DNA, and a nuclease that makes single-stranded nicks in DNA. It catalyses exonucleolytic cleavage in either 5′- to 3′- or 3′- to 5′-direction to yield 5′-phosphooligonucleotides.
RecQ helicase is a family of helicase enzymes initially found in Escherichia coli that has been shown to be important in genome maintenance. They function through catalyzing the reaction ATP + H2O → ADP + P and thus driving the unwinding of paired DNA and translocating in the 3' to 5' direction. These enzymes can also drive the reaction NTP + H2O → NDP + P to drive the unwinding of either DNA or RNA.

Non-homologous end joining (NHEJ) is a pathway that repairs double-strand breaks in DNA. It is called "non-homologous" because the break ends are directly ligated without the need for a homologous template, in contrast to homology directed repair (HDR), which requires a homologous sequence to guide repair. NHEJ is active in both non-dividing and proliferating cells, while HDR is not readily accessible in non-dividing cells. The term "non-homologous end joining" was coined in 1996 by Moore and Haber.

RecA is a 38 kilodalton protein essential for the repair and maintenance of DNA in bacteria. Structural and functional homologs to RecA have been found in all kingdoms of life. RecA serves as an archetype for this class of homologous DNA repair proteins. The homologous protein is called RAD51 in eukaryotes and RadA in archaea.

Nucleoproteins are proteins conjugated with nucleic acids. Typical nucleoproteins include ribosomes, nucleosomes and viral nucleocapsid proteins.

Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids.
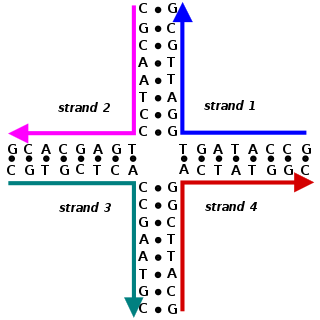
A Holliday junction is a branched nucleic acid structure that contains four double-stranded arms joined. These arms may adopt one of several conformations depending on buffer salt concentrations and the sequence of nucleobases closest to the junction. The structure is named after Robin Holliday, the molecular biologist who proposed its existence in 1964.
Recombinases are genetic recombination enzymes.
DNA repair protein RAD51 homolog 1 is a protein encoded by the gene RAD51. The enzyme encoded by this gene is a member of the RAD51 protein family which assists in repair of DNA double strand breaks. RAD51 family members are homologous to the bacterial RecA, Archaeal RadA, and yeast Rad51. The protein is highly conserved in most eukaryotes, from yeast to humans.

Spo11 is a protein that in humans is encoded by the SPO11 gene. Spo11, in a complex with mTopVIB, creates double strand breaks to initiate meiotic recombination. Its active site contains a tyrosine which ligates and dissociates with DNA to promote break formation. One Spo11 protein is involved per strand of DNA, thus two Spo11 proteins are involved in each double stranded break event.

Cell cycle checkpoint protein RAD17 is a protein that in humans is encoded by the RAD17 gene.

Bloom syndrome protein is a protein that in humans is encoded by the BLM gene and is not expressed in Bloom syndrome.

Replication protein A (RPA) is the major protein that binds to single-stranded DNA (ssDNA) in eukaryotic cells. In vitro, RPA shows a much higher affinity for ssDNA than RNA or double-stranded DNA. RPA is required in replication, recombination and repair processes such as nucleotide excision repair and homologous recombination. It also plays roles in responding to damaged DNA.

Cell cycle checkpoint protein RAD1 is a protein that in humans is encoded by the RAD1 gene.

Meiotic recombination protein DMC1/LIM15 homolog is a protein that in humans is encoded by the DMC1 gene.

Branch migration is the process by which base pairs on homologous DNA strands are consecutively exchanged at a Holliday junction, moving the branch point up or down the DNA sequence. Branch migration is the second step of genetic recombination, following the exchange of two single strands of DNA between two homologous chromosomes. The process is random, and the branch point can be displaced in either direction on the strand, influencing the degree of which the genetic material is exchanged. Branch migration can also be seen in DNA repair and replication, when filling in gaps in the sequence. It can also be seen when a foreign piece of DNA invades the strand.
Sgs1, also known as slow growth suppressor 1, is a DNA helicase protein found in Saccharomyces cerevisiae. It is a homolog of the bacterial RecQ helicase. Like the other members of the RecQ helicase family, Sgs1 is important for DNA repair. In particular, Sgs1 collaborates with other proteins to repair double-strand breaks during homologous recombination in eukaryotes.
The RecF pathway, also called the RecFOR pathway, is a pathway of homologous recombination that repairs DNA in bacteria. It repairs breaks that occur on only one of DNA's two strands, known as single-strand gaps. The RecF pathway can also repair double-strand breaks in DNA when the RecBCD pathway, another pathway of homologous recombination in bacteria, is inactivated by mutations. Like the RecBCD pathway, the RecF pathway requires RecA for strand invasion. The two pathways are also similar in their phases of branch migration, in which the Holliday junction slides in one direction, and resolution, in which the Holliday junctions are cleaved apart by enzymes.

Synthesis-dependent strand annealing (SDSA) is a major mechanism of homology-directed repair of DNA double-strand breaks (DSBs). Although many of the features of SDSA were first suggested in 1976, the double-Holliday junction model proposed in 1983 was favored by many researchers. In 1994, studies of double-strand gap repair in Drosophila were found to be incompatible with the double-Holliday junction model, leading researchers to propose a model they called synthesis-dependent strand annealing. Subsequent studies of meiotic recombination in S. cerevisiae found that non-crossover products appear earlier than double-Holliday junctions or crossover products, challenging the previous notion that both crossover and non-crossover products are produced by double-Holliday junctions and leading the authors to propose that non-crossover products are generated through SDSA.
References
- ↑ Seitz EM, Brockman JP, Sandler SJ, Clark AJ, Kowalczykowski SC (May 1998). "RadA protein is an archaeal RecA protein homolog that catalyzes DNA strand exchange". Genes & Development. 12 (9): 1248–53. doi:10.1101/gad.12.9.1248. PMC 316774 . PMID 9573041.