Related Research Articles

A retrovirus is a type of RNA virus that inserts a copy of its genome into the DNA of a host cell that it invades, thus changing the genome of that cell. Once inside the host cell's cytoplasm, the virus uses its own reverse transcriptase enzyme to produce DNA from its RNA genome, the reverse of the usual pattern, thus retro (backwards). The new DNA is then incorporated into the host cell genome by an integrase enzyme, at which point the retroviral DNA is referred to as a provirus. The host cell then treats the viral DNA as part of its own genome, transcribing and translating the viral genes along with the cell's own genes, producing the proteins required to assemble new copies of the virus.
An RNA virus is a virus that has RNA as its genetic material. This nucleic acid is usually single-stranded RNA (ssRNA) but may be double-stranded RNA (dsRNA). Notable human diseases caused by RNA viruses include the common cold, influenza, SARS, COVID-19, hepatitis C, hepatitis E, West Nile fever, Ebola virus disease, rabies, polio and measles.

A reverse transcriptase (RT) is an enzyme used to generate complementary DNA (cDNA) from an RNA template, a process termed reverse transcription. Reverse transcriptases are used by retroviruses to replicate their genomes, by retrotransposon mobile genetic elements to proliferate within the host genome, by eukaryotic cells to extend the telomeres at the ends of their linear chromosomes, and by some non-retroviruses such as the hepatitis B virus, a member of the Hepadnaviridae, which are dsDNA-RT viruses.
Virus classification is the process of naming viruses and placing them into a taxonomic system. Similar to the classification systems used for cellular organisms, virus classification is the subject of ongoing debate and proposals. This is mainly due to the pseudo-living nature of viruses, which is to say they are non-living particles with some chemical characteristics similar to those of life, or non-cellular life. As such, they do not fit neatly into the established biological classification system in place for cellular organisms.
Metaviridae are a family of viruses which exist as Ty3-gypsy LTR retrotransposons in a eukaryotic host’s genome. They are closely related to retroviruses: Metaviridae share many genomic elements with retroviruses, including length, organization, and genes themselves. This includes genes that encode reverse transcriptase, integrase, and capsid proteins. The reverse transcriptase and integrase proteins are needed for the retrotransposon activity of the virus. In some cases, virus-like particles can be formed from capsid proteins.
The Pseudoviridae are a family of viruses, including the following genera:
Caulimoviridae is a family of viruses infecting plants. There are currently 68 species in this family, divided among 8 genera. Viruses belonging to the family Caulimoviridae are termed double-stranded DNA (dsDNA) reverse-transcribing viruses i.e. viruses that contain a reverse transcription stage in their replication cycle. This family contains all plant viruses with a dsDNA genome that have a reverse transcribing phase in its lifecycle.

dsDNA-RT viruses are the seventh group in the Baltimore virus classification. They are not considered DNA viruses, but, rather reverse transcribing viruses because they replicate through an RNA intermediate. It includes the families Hepadnaviridae and Caulimoviridae.
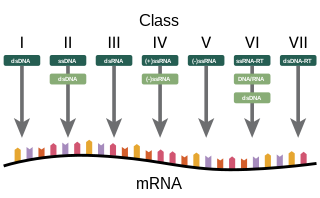
The Baltimore classification, developed by David Baltimore, is a virus classification system that groups viruses into families, depending on their type of genome and their method of replication.
Petuvirus is a genus of viruses, in the family Caulimoviridae order Ortervirales. Plants serve as natural hosts. There is currently only one species in this genus: the type species Petunia vein clearing virus. Diseases associated with this genus include: plants: chlorotic vein clearing, leaf malformation.
Avian sarcoma leukosis virus (ASLV) is an endogenous retrovirus that infects and can lead to cancer in chickens; experimentally it can infect other species of birds and mammals. ASLV replicates in chicken embryo fibroblasts, the cells that contribute to the formation of connective tissues. Different forms of the disease exist, including lymphoblastic, erythroblastic, and osteopetrotic.

LTR retrotransposons are class I transposable element characterized by the presence of Long Terminal Repeats (LTRs) directly flanking an internal coding region. As retrotransposons, they mobilize through reverse-transcription of their mRNA and integration of the newly created cDNA into another location. Their mechanism of retrotransposition is shared with retroviruses, with the difference that most LTR-retrotransposons do not form infectious particles that leave the cells and therefore only replicate inside their genome of origin. Those they do (occasionally) from virus-like particles are put under Ortervirales.
Badnavirus is a genus of viruses, in the family Caulimoviridae order Ortervirales. Plants serve as natural hosts. There are currently 32 species in this genus including the type species Commelina yellow mottle virus. Diseases associated with this genus include: CSSV: leaf chlorosis, root necrosis, red vein banding in young leaves, small mottled pods, and stem/root swelling followed by die-back. Infection decreases yield by 25% within one year, 50% within two years and usually kills trees within 3–4 years.
Caulimovirus is a genus of viruses, in the family Caulimoviridae order Ortervirales. Plants serve as natural hosts. There are currently ten species in this genus including the type species Cauliflower mosaic virus. Diseases associated with this genus include: vein-clearing or banding mosaic.
Cavemovirus is a genus of viruses, in the family Caulimoviridae order Ortervirales. Plants serve as natural hosts. There are currently only two species in this genus including the type species Cassava vein mosaic virus. Diseases associated with this genus include: vein-clearing or banding mosaic.
Soymovirus is a genus of viruses, in the family Caulimoviridae order Ortervirales. Plants serve as natural hosts. There are currently four species in this genus including the type species Soybean chlorotic mottle virus.
Tungrovirus is a genus of viruses, in the family Caulimoviridae order Ortervirales. Monocots and family Poaceae serve as natural hosts. There is currently only one species in this genus: the type species Rice tungro bacilliform virus. Diseases associated with this genus include: stunting, yellow to orange leaf discoloration with fewer tillers.
Solendovirus is a genus of viruses, in the family Caulimoviridae order Ortervirales. Plants serve as natural hosts. There are currently only two species in this genus, including the type species Tobacco vein clearing virus. Diseases associated with this genus include: TVCV: vein-clearing symptoms in N. edwardsonii.
Semotivirus is a genus of viruses in the family Belpaoviridae. Species exist as retrotransposons in a eukaryotic host's genome.
Ortervirales is an order that contains single-stranded RNA viruses that replicate through a DNA intermediate and double-stranded DNA viruses that replicate through an RNA intermediate . The name is derived from the reverse of retro.
References
- ↑ Krupovic, M; Blomberg, J; Coffin, JM; Dasgupta, I; Fan, H; Geering, AD; Gifford, R; Harrach, B; Hull, R; Johnson, W; Kreuze, JF; Lindemann, D; Llorens, C; Lockhart, B; Mayer, J; Muller, E; Olszewski, N; Pappu, HR; Pooggin, M; Richert-Pöggeler, KR; Sabanadzovic, S; Sanfaçon, H; Schoelz, JE; Seal, S; Stavolone, L; Stoye, JP; Teycheney, PY; Tristem, M; Koonin, EV; Kuhn, JH (4 April 2018). "Ortervirales: A new viral order unifying five families of reverse-transcribing viruses". Journal of Virology. 92 (12). doi:10.1128/JVI.00515-18. PMC 5974489 . PMID 29618642.
| This article includes a list of related items that share the same name (or similar names). If an internal link incorrectly led you here, you may wish to change the link to point directly to the intended article. |