Bioleaching is the extraction or liberation of metals from their ores through the use of living organisms. Bioleaching is one of several applications within biohydrometallurgy and several methods are used to treat ores or concentrates containing copper, zinc, lead, arsenic, antimony, nickel, molybdenum, gold, silver, and cobalt.

Oxidative phosphorylation or electron transport-linked phosphorylation or terminal oxidation is the metabolic pathway in which cells use enzymes to oxidize nutrients, thereby releasing chemical energy in order to produce adenosine triphosphate (ATP). In eukaryotes, this takes place inside mitochondria. Almost all aerobic organisms carry out oxidative phosphorylation. This pathway is so pervasive because it releases more energy than alternative fermentation processes such as anaerobic glycolysis.
An electron transport chain (ETC) is a series of protein complexes and other molecules that transfer electrons from electron donors to electron acceptors via redox reactions (both reduction and oxidation occurring simultaneously) and couples this electron transfer with the transfer of protons (H+ ions) across a membrane. Many of the enzymes in the electron transport chain are embedded within the membrane.

The cytochrome complex, or cyt c, is a small hemeprotein found loosely associated with the inner membrane of the mitochondrion where it plays a critical role in cellular respiration. It transfers electrons between Complexes III and IV. Cytochrome c is highly water-soluble, unlike other cytochromes. It is capable of undergoing oxidation and reduction as its iron atom converts between the ferrous and ferric forms, but does not bind oxygen. It also plays a major role in cell apoptosis. In humans, cytochrome c is encoded by the CYCS gene.

The enzyme cytochrome c oxidase or Complex IV, is a large transmembrane protein complex found in bacteria, archaea, and the mitochondria of eukaryotes.

Heme, or haem, is a ring-shaped iron-containing molecular component of hemoglobin, which is necessary to bind oxygen in the bloodstream. It is composed of four pyrrole rings with 2 vinyl and 2 propionic acid side chains. Heme is biosynthesized in both the bone marrow and the liver.

Acidithiobacillus is a genus of the Acidithiobacillia in the phylum "Pseudomonadota". This genus includes ten species of acidophilic microorganisms capable of sulfur and/or iron oxidation: Acidithiobacillus albertensis, Acidithiobacillus caldus, Acidithiobacillus cuprithermicus, Acidithiobacillus ferrianus, Acidithiobacillus ferridurans, Acidithiobacillus ferriphilus, Acidithiobacillus ferrivorans, Acidithiobacillus ferrooxidans, Acidithiobacillus sulfuriphilus, and Acidithiobacillus thiooxidans.A. ferooxidans is the most widely studied of the genus, but A. caldus and A. thiooxidans are also significant in research. Like all "Pseudomonadota", Acidithiobacillus spp. are Gram-negative and non-spore forming. They also play a significant role in the generation of acid mine drainage; a major global environmental challenge within the mining industry. Some species of Acidithiobacillus are utilized in bioleaching and biomining. A portion of the genes that support the survival of these bacteria in acidic environments are presumed to have been obtained by horizontal gene transfer.
Ferroxidase also known as Fe(II):oxygen oxidoreductase is an enzyme that catalyzes the oxidization of iron II to iron III:
Microbial metabolism is the means by which a microbe obtains the energy and nutrients it needs to live and reproduce. Microbes use many different types of metabolic strategies and species can often be differentiated from each other based on metabolic characteristics. The specific metabolic properties of a microbe are the major factors in determining that microbe's ecological niche, and often allow for that microbe to be useful in industrial processes or responsible for biogeochemical cycles.
Biomining refers to any process that uses living organisms to extract metals from ores and other solid materials. Typically these processes involve prokaryotes, however fungi and plants may also be used. Biomining is one of several applications within biohydrometallurgy with applications in ore refinement, precious metal recovery, and bioremediation. The largest application currently being used is the treatment of mining waste containing iron, copper, zinc, and gold allowing for salvation of any discarded minerals. It may also be useful in maximizing the yields of increasingly low grade ore deposits. Biomining has been proposed as a relatively environmentally friendly alternative and/or supplementation to traditional mining. Current methods of biomining are modified leach mining processes. These aptly named bioleaching processes most commonly includes the inoculation of extracted rock with bacteria and acidic solution, with the leachate salvaged and processed for the metals of value. Biomining has many applications outside of metal recovery, most notably is bioremediation which has already been used to clean up coastlines after oil spills. There are also many promising future applications, like space biomining, fungal bioleaching and biomining with hybrid biomaterials.
Nitrite oxidoreductase is an enzyme involved in nitrification. It is the last step in the process of aerobic ammonia oxidation, which is carried out by two groups of nitrifying bacteria: ammonia oxidizers such as Nitrosospira, Nitrosomonas, and Nitrosococcus convert ammonia to nitrite, while nitrite oxidizers such as Nitrobacter and Nitrospira oxidize nitrite to nitrate. NXR is responsible for producing almost all nitrate found in nature.

Thiosulfate dehydrogenase is an enzyme that catalyzes the chemical reaction:
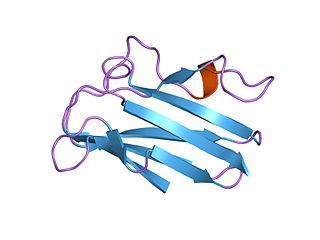
Plastocyanin/azurin family of copper-binding proteins is a family of small proteins that bind a single copper atom and that are characterised by an intense electronic absorption band near 600 nm. The most well-known members of this class of proteins are the plant chloroplastic plastocyanins, which exchange electrons with cytochrome c6, and the distantly related bacterial azurins, which exchange electrons with cytochrome c551. This family of proteins also includes amicyanin from bacteria such as Methylobacterium extorquens or Paracoccus versutus that can grow on methylamine; auracyanins A and B from Chloroflexus aurantiacus; blue copper protein from Alcaligenes faecalis; cupredoxin (CPC) from Cucumis sativus (Cucumber) peelings; cusacyanin from cucumber; halocyanin from Natronomonas pharaonis, a membrane-associated copper-binding protein; pseudoazurin from Pseudomonas; rusticyanin from Thiobacillus ferrooxidans; stellacyanin from Rhus vernicifera ; umecyanin from the roots of Armoracia rusticana (Horseradish); and allergen Ra3 from ragweed. This pollen protein has evolutary relation to the above proteins, but seems to have lost the ability to bind copper. Although there is an appreciable amount of divergence in the sequences of all these proteins, the copper ligand sites are conserved.

Cytochrome c oxidase subunit 4 isoform 2, mitochondrial is an enzyme that in humans is encoded by the COX4I2 gene. COX4I2 is a nuclear-encoded isoform of cytochrome c oxidase (COX) subunit 4. Cytochrome c oxidase is a multi-subunit enzyme complex that couples the transfer of electrons from cytochrome c to molecular oxygen and contributes to a proton electrochemical gradient across the inner mitochondrial membrane, acting as the terminal enzyme of the mitochondrial respiratory chain. Mutations in COX4I2 have been associated with exocrine pancreatic insufficiency, dyserythropoietic anemia, and calvarial hyperostosis (EPIDACH).

The outflow of acidic liquids and other pollutants from mines is often catalysed by acid-loving microorganisms; these are the acidophiles in acid mine drainage.
The Arc system is a two-component system found in some bacteria that regulates gene expression in faculatative anaerobes such as Escheria coli. Two-component system means that it has a sensor molecule and a response regulator. Arc is an abbreviation for Anoxic Redox Control system. Arc systems are instrumental in maintaining energy metabolism during transcription of bacteria. The ArcA response regulator looks at growth conditions and expresses genes to best suit the bacteria. The Arc B sensor kinase, which is a tripartite protein, is membrane bound and can autophosphorylate.
Sulfide:quinone reductase is an enzyme with systematic name sulfide:quinone oxidoreductase. This enzyme catalyses the following chemical reaction
Iron:rusticyanin reductase is an enzyme with systematic name Fe(II):rusticyanin oxidoreductase. This enzyme catalyses the following chemical reaction
Acidithiobacillus caldus formerly belonged to the genus Thiobacillus prior to 2000, when it was reclassified along with a number of other bacterial species into one of three new genera that better categorize sulfur-oxidizing acidophiles. As a member of the Gammaproteobacteria class of Pseudomonadota, A. caldus may be identified as a Gram-negative bacterium that is frequently found in pairs. Considered to be one of the most common microbes involved in biomining, it is capable of oxidizing reduced inorganic sulfur compounds (RISCs) that form during the breakdown of sulfide minerals. The meaning of the prefix acidi- in the name Acidithiobacillus comes from the Latin word acidus, signifying that members of this genus love a sour, acidic environment. Thio is derived from the Greek word thios and describes the use of sulfur as an energy source, and bacillus describes the shape of these microorganisms, which are small rods. The species name, caldus, is derived from the Latin word for warm or hot, denoting this species' love of a warm environment.
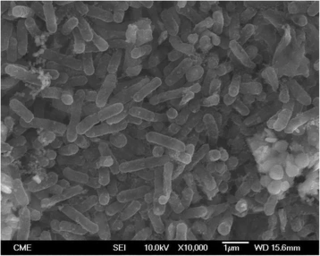
Acidithiobacillus thiooxidans, formerly known as Thiobacillus thiooxidans until its reclassification into the newly designated genus Acidithiobacillus of the Acidithiobacillia subclass of Pseudomonadota, is a Gram-negative, rod-shaped bacterium that uses sulfur as its primary energy source. It is mesophilic, with a temperature optimum of 28 °C. This bacterium is commonly found in soil, sewer pipes, and cave biofilms called snottites. A. thiooxidans is used in the mining technique known as bioleaching, where metals are extracted from their ores through the action of microbes.










