
An extremophile is an organism that is able to live in extreme environments, i.e., environments with conditions approaching or stretching the limits of what known life can adapt to, such as extreme temperature, pressure, radiation, salinity, or pH level.

Metagenomics is the study of genetic material recovered directly from environmental or clinical samples by a method called sequencing. The broad field may also be referred to as environmental genomics, ecogenomics, community genomics or microbiomics.

Acidithiobacillus is a genus of the Acidithiobacillia in the phylum "Pseudomonadota". This genus includes ten species of acidophilic microorganisms capable of sulfur and/or iron oxidation: Acidithiobacillus albertensis, Acidithiobacillus caldus, Acidithiobacillus cuprithermicus, Acidithiobacillus ferrianus, Acidithiobacillus ferridurans, Acidithiobacillus ferriphilus, Acidithiobacillus ferrivorans, Acidithiobacillus ferrooxidans, Acidithiobacillus sulfuriphilus, and Acidithiobacillus thiooxidans.A. ferooxidans is the most widely studied of the genus, but A. caldus and A. thiooxidans are also significant in research. Like all "Pseudomonadota", Acidithiobacillus spp. are Gram-negative and non-spore forming. They also play a significant role in the generation of acid mine drainage; a major global environmental challenge within the mining industry. Some species of Acidithiobacillus are utilized in bioleaching and biomining. A portion of the genes that support the survival of these bacteria in acidic environments are presumed to have been obtained by horizontal gene transfer.

Sulfur-reducing bacteria are microorganisms able to reduce elemental sulfur (S0) to hydrogen sulfide (H2S). These microbes use inorganic sulfur compounds as electron acceptors to sustain several activities such as respiration, conserving energy and growth, in absence of oxygen. The final product of these processes, sulfide, has a considerable influence on the chemistry of the environment and, in addition, is used as electron donor for a large variety of microbial metabolisms. Several types of bacteria and many non-methanogenic archaea can reduce sulfur. Microbial sulfur reduction was already shown in early studies, which highlighted the first proof of S0 reduction in a vibrioid bacterium from mud, with sulfur as electron acceptor and H
2 as electron donor. The first pure cultured species of sulfur-reducing bacteria, Desulfuromonas acetoxidans, was discovered in 1976 and described by Pfennig Norbert and Biebel Hanno as an anaerobic sulfur-reducing and acetate-oxidizing bacterium, not able to reduce sulfate. Only few taxa are true sulfur-reducing bacteria, using sulfur reduction as the only or main catabolic reaction. Normally, they couple this reaction with the oxidation of acetate, succinate or other organic compounds. In general, sulfate-reducing bacteria are able to use both sulfate and elemental sulfur as electron acceptors. Thanks to its abundancy and thermodynamic stability, sulfate is the most studied electron acceptor for anaerobic respiration that involves sulfur compounds. Elemental sulfur, however, is very abundant and important, especially in deep-sea hydrothermal vents, hot springs and other extreme environments, making its isolation more difficult. Some bacteria – such as Proteus, Campylobacter, Pseudomonas and Salmonella – have the ability to reduce sulfur, but can also use oxygen and other terminal electron acceptors.

Myxococcus xanthus is a gram-negative, bacillus species of myxobacteria that is typically found in the top-most layer of soil. These bacteria lack flagella; rather, they use pili for motility. M. xanthus is well-known for its predatory behavior on other microorganisms. These bacteria source carbon from lipids rather than sugars. They exhibit various forms of self-organizing behavior in response to environmental cues. Under normal conditions with abundant food, they exist as predatory, saprophytic single-species biofilm called a swarm, highlighting the importance of intercellular communication for these bacteria. Under starvation conditions, they undergo a multicellular development cycle.
Biomining refers to any process that uses living organisms to extract metals from ores and other solid materials. Typically these processes involve prokaryotes, however fungi and plants may also be used. Biomining is one of several applications within biohydrometallurgy with applications in ore refinement, precious metal recovery, and bioremediation. The largest application currently being used is the treatment of mining waste containing iron, copper, zinc, and gold allowing for salvation of any discarded minerals. It may also be useful in maximizing the yields of increasingly low grade ore deposits. Biomining has been proposed as a relatively environmentally friendly alternative and/or supplementation to traditional mining. Current methods of biomining are modified leach mining processes. These aptly named bioleaching processes most commonly includes the inoculation of extracted rock with bacteria and acidic solution, with the leachate salvaged and processed for the metals of value. Biomining has many applications outside of metal recovery, most notably is bioremediation which has already been used to clean up coastlines after oil spills. There are also many promising future applications, like space biomining, fungal bioleaching and biomining with hybrid biomaterials.

The outflow of acidic liquids and other pollutants from mines is often catalysed by acid-loving microorganisms; these are the acidophiles in acid mine drainage.

In the fields of molecular biology and genetics, a pan-genome is the entire set of genes from all strains within a clade. More generally, it is the union of all the genomes of a clade. The pan-genome can be broken down into a "core pangenome" that contains genes present in all individuals, a "shell pangenome" that contains genes present in two or more strains, and a "cloud pangenome" that contains genes only found in a single strain. Some authors also refer to the cloud genome as "accessory genome" containing 'dispensable' genes present in a subset of the strains and strain-specific genes. Note that the use of the term 'dispensable' has been questioned, at least in plant genomes, as accessory genes play "an important role in genome evolution and in the complex interplay between the genome and the environment". The field of study of pangenomes is called pangenomics.
Staphylococcus carnosus is a bacterium from the genus Staphylococcus that is both Gram-positive and coagulase-negative. It was originally identified in dry sausage and is an important starter culture for meat fermentation. Unlike other members of its genus, such as Staphylococcus aureus and Staphylococcus epidermidis, S. carnosus is nonpathogenic and safely used in the food industry.
Iron:rusticyanin reductase is an enzyme with systematic name Fe(II):rusticyanin oxidoreductase. This enzyme catalyses the following chemical reaction
Leptospirillum ferriphilum is an iron-oxidising bacterium able to exist in environments of high acidity, high iron concentrations, and moderate to moderately high temperatures. It is one of the species responsible for the generation of acid mine drainage and the principal microbe used in industrial biohydrometallurgy processes to extract metals.
Spiroplasma citri is a bacterium species and the causative agent of Citrus stubborn disease.
Acidithiobacillus caldus formerly belonged to the genus Thiobacillus prior to 2000, when it was reclassified along with a number of other bacterial species into one of three new genera that better categorize sulfur-oxidizing acidophiles. As a member of the Gammaproteobacteria class of Pseudomonadota, A. caldus may be identified as a Gram-negative bacterium that is frequently found in pairs. Considered to be one of the most common microbes involved in biomining, it is capable of oxidizing reduced inorganic sulfur compounds (RISCs) that form during the breakdown of sulfide minerals. The meaning of the prefix acidi- in the name Acidithiobacillus comes from the Latin word acidus, signifying that members of this genus love a sour, acidic environment. Thio is derived from the Greek word thios and describes the use of sulfur as an energy source, and bacillus describes the shape of these microorganisms, which are small rods. The species name, caldus, is derived from the Latin word for warm or hot, denoting this species' love of a warm environment.
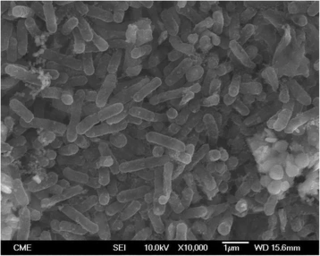
Acidithiobacillus thiooxidans, formerly known as Thiobacillus thiooxidans until its reclassification into the newly designated genus Acidithiobacillus of the Acidithiobacillia subclass of Pseudomonadota, is a Gram-negative, rod-shaped bacterium that uses sulfur as its primary energy source. It is mesophilic, with a temperature optimum of 28 °C. This bacterium is commonly found in soil, sewer pipes, and cave biofilms called snottites. A. thiooxidans is used in the mining technique known as bioleaching, where metals are extracted from their ores through the action of microbes.

ParaHoxozoa is a clade of animals that consists of Bilateria, Placozoa, and Cnidaria.

Corynebacterium glutamicum is a Gram-positive, rod-shaped bacterium that is used industrially for large-scale production of amino acids, especially glutamic acid and lysine. While originally identified in a screen for organisms secreting L-glutamate, mutants of C. glutamicum have also been identified that produce various other amino acids and derivatives of amino acids.

Salinibacter ruber is an extremely halophilic red bacterium, first found in Spain in 2002.

Shiladitya DasSarma is a molecular biologist well-known for contributions to the biology of halophilic and extremophilic microorganisms. He is a Professor in the University of Maryland Baltimore. He earned a PhD degree in biochemistry from the Massachusetts Institute of Technology and a BS degree in chemistry from Indiana University Bloomington. Prior to taking a faculty position, he conducted research at the Massachusetts General Hospital, Harvard Medical School, and Pasteur Institute, Paris.
Acidithrix ferrooxidans is a heterotrophic, acidophilic and Gram-positive bacterium from the genus Acidithrix. The type strain of this species, A. ferrooxidans Py-F3, was isolated from an acidic stream draining from a copper mine in Wales. This species grows in a variety of acidic environments such as streams, mines or geothermal sites. Mine lakes with a redoxcline support growth with ferrous iron as the electron donor. "A. ferrooxidans" grows rapidly in macroscopic streamer, producing greater cell densities than other streamer-forming microbes. Use in a bioreactors to remediate mine waste has been proposed due to cell densities and rapid oxidation of ferrous iron oxidation in acidic mine drainage. Exopolysaccharide production during metal substrate metabolism, such as iron oxidation helps to prevent cell encrustation by minerals.

Sulfobacillus is a genus of bacteria containing six named species. Members of the genus are Gram-positive, acidophilic, spore-forming bacteria that are moderately thermophilic or thermotolerant. All species are facultative anaerobes capable of oxidizing sulfur-containing compounds; they differ in optimal growth temperature and metabolic capacity, particularly in their ability to grow on various organic carbon compounds.













