Related Research Articles

A molecular lesion or point lesion is damage to the structure of a biological molecule such as DNA, RNA, or protein. This damage may result in the reduction or absence of normal function, and in rare cases the gain of a new function. Lesions in DNA may consist of breaks or other changes in chemical structure of the helix, ultimately preventing transcription. Meanwhile, lesions in proteins consist of both broken bonds and improper folding of the amino acid chain. While many nucleic acid lesions are general across DNA and RNA, some are specific to one, such as thymine dimers being found exclusively in DNA. Several cellular repair mechanisms exist, ranging from global to specific, in order to prevent lasting damage resulting from lesions.
DNA glycosylases are a family of enzymes involved in base excision repair, classified under EC number EC 3.2.2. Base excision repair is the mechanism by which damaged bases in DNA are removed and replaced. DNA glycosylases catalyze the first step of this process. They remove the damaged nitrogenous base while leaving the sugar-phosphate backbone intact, creating an apurinic/apyrimidinic site, commonly referred to as an AP site. This is accomplished by flipping the damaged base out of the double helix followed by cleavage of the N-glycosidic bond.

Base excision repair (BER) is a cellular mechanism, studied in the fields of biochemistry and genetics, that repairs damaged DNA throughout the cell cycle. It is responsible primarily for removing small, non-helix-distorting base lesions from the genome. The related nucleotide excision repair pathway repairs bulky helix-distorting lesions. BER is important for removing damaged bases that could otherwise cause mutations by mispairing or lead to breaks in DNA during replication. BER is initiated by DNA glycosylases, which recognize and remove specific damaged or inappropriate bases, forming AP sites. These are then cleaved by an AP endonuclease. The resulting single-strand break can then be processed by either short-patch or long-patch BER.

MUTYH is a human gene that encodes a DNA glycosylase, MUTYH glycosylase. It is involved in oxidative DNA damage repair and is part of the base excision repair pathway. The enzyme excises adenine bases from the DNA backbone at sites where adenine is inappropriately paired with guanine, cytosine, or 8-oxo-7,8-dihydroguanine, a common form of oxidative DNA damage.
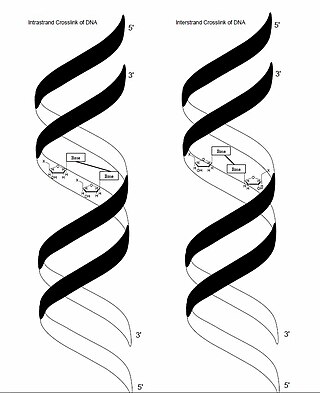
In genetics, crosslinking of DNA occurs when various exogenous or endogenous agents react with two nucleotides of DNA, forming a covalent linkage between them. This crosslink can occur within the same strand (intrastrand) or between opposite strands of double-stranded DNA (interstrand). These adducts interfere with cellular metabolism, such as DNA replication and transcription, triggering cell death. These crosslinks can, however, be repaired through excision or recombination pathways.

8-Oxoguanine glycosylase, also known as OGG1, is a DNA glycosylase enzyme that, in humans, is encoded by the OGG1 gene. It is involved in base excision repair. It is found in bacterial, archaeal and eukaryotic species.

Evelyn M. Witkin was an American bacterial geneticist at Cold Spring Harbor Laboratory (1944–1955), SUNY Downstate Medical Center (1955–1971), and Rutgers University (1971–1991). Witkin was considered innovative and inspirational as a scientist, teacher and mentor.

Cell cycle checkpoint control protein RAD9A is a protein that in humans is encoded by the RAD9A gene.Rad9 has been shown to induce G2 arrest in the cell cycle in response to DNA damage in yeast cells. Rad9 was originally found in budding yeast cells but a human homolog has also been found and studies have suggested that the molecular mechanisms of the S and G2 checkpoints are conserved in eukaryotes. Thus, what is found in yeast cells are likely to be similar in human cells.

UV excision repair protein RAD23 homolog B is a protein that in humans is encoded by the RAD23B gene.
Uracil-DNA glycosylase is an enzyme. Its most important function is to prevent mutagenesis by eliminating uracil from DNA molecules by cleaving the N-glycosidic bond and initiating the base-excision repair (BER) pathway.

Cyclin-O is a protein that in humans is encoded by the CCNO gene.

DNA-3-methyladenine glycosylase also known as 3-alkyladenine DNA glycosylase (AAG) or N-methylpurine DNA glycosylase (MPG) is an enzyme that in humans is encoded by the MPG gene.
DNA damage-binding protein or UV-DDB is a protein complex that is responsible for repair of UV-damaged DNA. This complex is composed of two protein subunits, a large subunit DDB1 (p127) and a small subunit DDB2 (p48). When cells are exposed to UV radiation, DDB1 moves from the cytosol to the nucleus and binds to DDB2, thus forming the UV-DDB complex. This complex formation is highly favorable and it is demonstrated by UV-DDB's binding preference and high affinity to the UV lesions in the DNA. This complex functions in nucleotide excision repair, recognising UV-induced (6-4) pyrimidine-pyrimidone photoproducts and cyclobutane pyrimidine dimers.

In molecular biology, the H2TH domain is a DNA-binding domain found in DNA glycosylase/AP lyase enzymes, which are involved in base excision repair of DNA damaged by oxidation or by mutagenic agents. Most damage to bases in DNA is repaired by the base excision repair pathway. These enzymes are primarily from bacteria, and have both DNA glycosylase activity EC 3.2.2.- and AP lyase activity EC 4.2.99.18. Examples include formamidopyrimidine-DNA glycosylases and endonuclease VIII (Nei).
DNA-deoxyinosine glycosylase is an enzyme with systematic name DNA-deoxyinosine deoxyribohydrolase. This enzyme is involved in DNA damage repair and targets hypoxanthine bases.

Leona D. Samson is the Uncas and Helen Whitaker Professor and American Cancer Society Research Professor of Biological Engineering at the Massachusetts Institute of Technology, where she served as the Director of the Center for Environmental Health Sciences from 2001 to 2012. Before her professorship at MIT, she held a professorship at the Harvard School of Public Health. She is on the editorial board of the journal DNA Repair. Her research interests focus on "methods for measuring DNA repair capacity (DRC) in human cells", research the National Institute of Health recognized as pioneering in her field, for which the NIH granted her the National Institutes of Health Director's Pioneer Award.

AlkD is an enzyme belonging to a family of DNA glycosylases that are involved in DNA repair. It was discovered by a team of Norwegian biologists from Oslo in 2006. It was isolated from a soil-dwelling Gram-positive bacteria Bacillus cereus, along with another enzyme AlkC. AlkC and AlkD are most probably derived from the same protein as indicated by their close resemblance. They are also found in other prokaryotes. Among eukaryotes, they are found only in the single-celled species only, such as Entamoeba histolytica and Dictyostelium discoideum. The enzyme specifically targets 7mG (methyl-guanine) in the DNA, and is, therefore, unique among DNA glycosylases. It can also act on other methylpurines with less affinity. It indicates that the enzyme is specific for locating and cutting (excision) of chemically modified bases from DNA, exactly at 7mG, whenever there are errors in replication. It accelerates the rate of 7mG hydrolysis 100-fold over the spontaneous depurination. Thus, it protects the genome from harmful changes induced by chemical and environmental agents. Its crystal structure was described in 2008. It is the first HEAT repeat protein identified to interact with nucleic acids or to contain enzymatic activity.

Cynthia J. Burrows is an American chemist, currently a distinguished professor in the department of chemistry at the University of Utah, where she is also the Thatcher Presidential Endowed Chair of Biological Chemistry. Burrows was the Senior Editor of the Journal of Organic Chemistry (2001-2013) and became Editor-in-Chief of Accounts of Chemical Research in 2014.,,

Orlando David Schärer is a Swiss chemist and biologist researching DNA repair, genomic integrity, and cancer biology. Schärer has taught biology, chemistry and pharmacology at various university levels on three continents. He is a distinguished professor at the Ulsan National Institute of Science and Technology (UNIST) and the associate director of the IBS Center for Genomic Integrity located in Ulsan, South Korea. He leads the three interdisciplinary research teams in the Chemical & Cancer Biology Branch of the center and specifically heads the Cancer Therapeutics Mechanisms Section.
Suse Broyde is an American chemical biologist who is a professor at New York University. Her research considers the mechanisms that underpin DNA damage. Broyde is the author of the Wiley textbook The Chemical Biology of DNA Damage.
References
- ↑ David, Sheila Sue (1989). "Spectroscopic studies of inhibitor binding to uteroferrin".
- ↑ Graduate School Commencement Program. University of Minnesota. Spring 1990. p. 12. hdl:11299/154961.
- ↑ "Group Members". The David Lab. Retrieved 2023-09-18.
- 1 2 "Research". The David Lab. Retrieved 2023-09-19.
- ↑ "2011 ACS Fellows". Chemical & Engineering News. Retrieved 2023-09-18.
- ↑ "AAAS Members Elected as Fellows". 2011-01-11.
- ↑ "Sheila S. David - Editorial Board - DNA Repair - Journal - Elsevier". www.journals.elsevier.com. Retrieved 2023-09-19.
- ↑ Alebee13 (2023-02-02). "Congratulations to Professor Sheila David on Receiving the 2022 Education Award". The David Lab. Retrieved 2023-09-18.