Related Research Articles
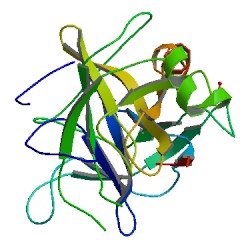
Chymotrypsin (EC 3.4.21.1, chymotrypsins A and B, alpha-chymar ophth, avazyme, chymar, chymotest, enzeon, quimar, quimotrase, alpha-chymar, alpha-chymotrypsin A, alpha-chymotrypsin) is a digestive enzyme component of pancreatic juice acting in the duodenum, where it performs proteolysis, the breakdown of proteins and polypeptides. Chymotrypsin preferentially cleaves peptide amide bonds where the side chain of the amino acid N-terminal to the scissile amide bond (the P1 position) is a large hydrophobic amino acid (tyrosine, tryptophan, and phenylalanine). These amino acids contain an aromatic ring in their side chain that fits into a hydrophobic pocket (the S1 position) of the enzyme. It is activated in the presence of trypsin. The hydrophobic and shape complementarity between the peptide substrate P1 side chain and the enzyme S1 binding cavity accounts for the substrate specificity of this enzyme. Chymotrypsin also hydrolyzes other amide bonds in peptides at slower rates, particularly those containing leucine at the P1 position.

Protein folding is the physical process by which a protein chain is translated into its native three-dimensional structure, typically a "folded" conformation, by which the protein becomes biologically functional. Via an expeditious and reproducible process, a polypeptide folds into its characteristic three-dimensional structure from a random coil. Each protein exists first as an unfolded polypeptide or random coil after being translated from a sequence of mRNA into a linear chain of amino acids. At this stage, the polypeptide lacks any stable three-dimensional structure. As the polypeptide chain is being synthesized by a ribosome, the linear chain begins to fold into its three-dimensional structure.
Levinthal's paradox is a thought experiment, also constituting a self-reference in the theory of protein folding. In 1969, Cyrus Levinthal noted that, because of the very large number of degrees of freedom in an unfolded polypeptide chain, the molecule has an astronomical number of possible conformations. An estimate of 10300 was made in one of his papers (often incorrectly cited as the 1968 paper). For example, a polypeptide of 100 residues will have 99 peptide bonds, and therefore 198 different phi and psi bond angles. If each of these bond angles can be in one of three stable conformations, the protein may misfold into a maximum of 3198 different conformations (including any possible folding redundancy). Therefore, if a protein were to attain its correctly folded configuration by sequentially sampling all the possible conformations, it would require a time longer than the age of the universe to arrive at its correct native conformation. This is true even if conformations are sampled at rapid (nanosecond or picosecond) rates. The "paradox" is that most small proteins fold spontaneously on a millisecond or even microsecond time scale. The solution to this paradox has been established by computational approaches to protein structure prediction.

Sir Alan Roy Fersht is a British chemist at the MRC Laboratory of Molecular Biology, Cambridge, and an Emeritus Professor in the Department of Chemistry at the University of Cambridge. He was Master of Gonville and Caius College, Cambridge from 2012 to 2018. He works on protein folding, and is sometimes described as a founder of protein engineering.

In chemistry, a molecular knot is a mechanically interlocked molecular architecture that is analogous to a macroscopic knot. Naturally-forming molecular knots are found in organic molecules like DNA, RNA, and proteins. It is not certain that naturally occurring knots are evolutionarily advantageous to nucleic acids or proteins, though knotting is thought to play a role in the structure, stability, and function of knotted biological molecules. The mechanism by which knots naturally form in molecules, and the mechanism by which a molecule is stabilized or improved by knotting, is ambiguous. The study of molecular knots involves the formation and applications of both naturally occurring and chemically synthesized molecular knots. Applying chemical topology and knot theory to molecular knots allows biologists to better understand the structures and synthesis of knotted organic molecules.
Phi value analysis, analysis, or -value analysis is an experimental protein engineering technique for studying the structure of the folding transition state of small protein domains that fold in a two-state manner. The structure of the folding transition state is hard to find using methods such as protein NMR or X-ray crystallography because folding transitions states are mobile and partly unstructured by definition. In -value analysis, the folding kinetics and conformational folding stability of the wild-type protein are compared with those of point mutants to find phi values. These measure the mutant residue's energetic contribution to the folding transition state, which reveals the degree of native structure around the mutated residue in the transition state, by accounting for the relative free energies of the unfolded state, the folded state, and the transition state for the wild-type and mutant proteins.
Hydrophobic collapse is a proposed process for the production of the 3-D conformation adopted by polypeptides and other molecules in polar solvents. The theory states that the nascent polypeptide forms initial secondary structure creating localized regions of predominantly hydrophobic residues. The polypeptide interacts with water, thus placing thermodynamic pressures on these regions which then aggregate or "collapse" into a tertiary conformation with a hydrophobic core. Incidentally, polar residues interact favourably with water, thus the solvent-facing surface of the peptide is usually composed of predominantly hydrophilic regions.
Downhill folding is a process in which a protein folds without encountering any significant macroscopic free energy barrier. It is a key prediction of the folding funnel hypothesis of the energy landscape theory of proteins.
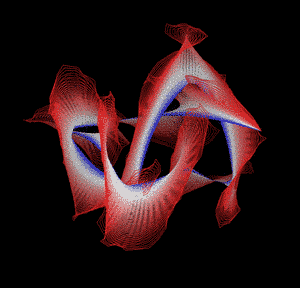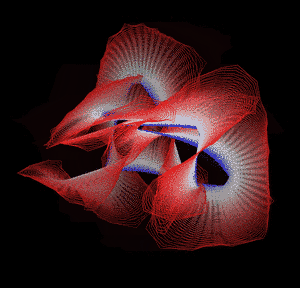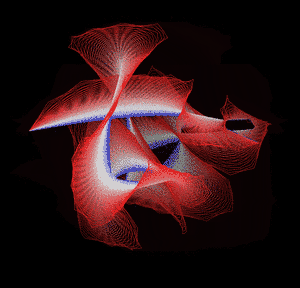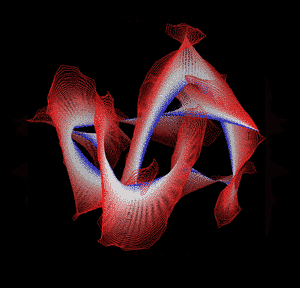
Knotted proteins are proteins whose backbones entangle themselves in a knot. One can imagine pulling a protein chain from both termini, as though pulling a string from both ends. When a knotted protein is “pulled” from both termini, it does not get disentangled. Knotted proteins are very rare, making up only about one percent of the proteins in the Protein Data Bank, and their folding mechanisms and function are not well understood. Although there are experimental and theoretical studies that hint to some answers, systematic answers to these questions have not yet been found.
Sheena Elizabeth Radford FRS FMedSci is a British biophysicist, and Astbury Professor of Biophysics in the Astbury Centre for Structural Molecular Biology, School of Molecular and Cellular Biology at the University of Leeds. Radford is the Associate Editor of the Journal of Molecular Biology.

Judith P. Klinman is an American chemist, biochemist, and molecular biologist known for her work on enzyme catalysis. She became the first female professor in the physical sciences at the University of California, Berkeley in 1978, where she is now Professor of the Graduate School and Chancellor's Professor. In 2012, she was awarded the National Medal of Science by President Barack Obama. She is a member of the National Academy of Sciences, American Academy of Arts and Sciences, American Association for the Advancement of Science, and the American Philosophical Society.

Jane Clarke is an English biochemist and academic. Since October 2017, she has served as President of Wolfson College, Cambridge. She is also Professor of Molecular Biophysics, a Wellcome Trust Senior Research Fellow in the Department of Chemistry at the University of Cambridge. She was previously a Fellow of Trinity Hall, Cambridge.
Brian Selby Hartley FRS was a British biochemist. He was Professor of Biochemistry at Imperial College London from 1974 to 1991.
Anne Bertolotti is a French biochemist and cell biologist who works as Programme Leader at the MRC Laboratory of Molecular Biology in Cambridge, UK. In 2022 she was appointed Head of the MRC LMB's Neurobiology Division. She is known for her research into the cellular defences against misfolded proteins and the mechanisms underlying their deposition, the molecular problem causative of neurodegenerative diseases.
Deborah Beth Zamble was a Canadian chemist and Canada Research Chair in Biological Chemistry at the University of Toronto. Her research considered how bacteria processed metal nutrients.
Jean Baum is an American chemist. She is the Distinguished Professor of Chemistry and Chemical Biology at Rutgers University, where she is also Vice Dean for Research and Graduate Education in the School of Arts and Sciences, and also Vice Chair of the Department of Chemistry and Chemical Biology. Her research investigates protein–protein interaction and protein aggregation using nuclear magnetic resonance spectroscopy (NMR) and other biochemical and biophysical techniques. She serves as Treasurer for the Protein Society.
Suzanne Frances Scarlata is the Richard Whitcomb Professor at Worcester Polytechnic Institute. She is known for her work on how cells respond to hormones and neurotransmitters. She is an elected fellow of the American Association for the Advancement of Science.
Edith Wilson Miles is a biochemist known for her work on the structure and function of enzymes, especially her work on tryptophan synthase.
Linda Columbus is an American chemist who is Professor of Chemistry and Molecular Physiology at the University of Virginia. Her research considers the structure-function properties of membrane proteins.
Elizabeth Sally Ward is a British physician who is Director of Translational Immunology at the Centre for Cancer Immunology in the University of Southampton. She was elected Fellow of the Royal Society in 2022.
References
- 1 2 3 4 5 6 7 8 "Sophie Jackson | Yusuf Hamied Department of Chemistry". www.ch.cam.ac.uk. Retrieved 2023-04-01.
- 1 2 3 4 "Biological self assembly: from self tying proteins to microcrystalline suspensions of peptides, Wednesday 9 February, 2:00pm". www.lancaster.ac.uk. Retrieved 2023-04-01.
- 1 2 3 "Jackson Group Sophies Page". www-jackson.ch.cam.ac.uk. Retrieved 2023-04-01.
- ↑ "Video: Sophie Jackson, "Protein Knots: Experimental Studies on Stability, Folding, Degradation and Design"". www.birs.ca. Retrieved 2023-04-01.
- 1 2 3 "The Mystery of Knotted Proteins". MIT Technology Review. Retrieved 2023-04-01.
- 1 2 3 Bennett2010-09-29T12:35:00+01:00, Hayley. "Protein folding: knotted or not". Chemistry World. Retrieved 2023-04-01.