Drosophila embryogenesis, the process by which Drosophila embryos form, is a favorite model system for genetics and developmental biology. The study of its embryogenesis unlocked the century-long puzzle of how development was controlled, creating the field of evolutionary developmental biology. The small size, short generation time, and large brood size make it ideal for genetic studies. Transparent embryos facilitate developmental studies. Drosophila melanogaster was introduced into the field of genetic experiments by Thomas Hunt Morgan in 1909.
The border cells are a cluster of 6–8 cells that migrate in the ovariole of the fruit-fly Drosophila melanogaster, during the process of oogenesis. A fly ovary consists of a string of ovarioles or egg chambers arranged in an increasing order of maturity. Each egg chamber contains 16 central germline, nurse cells surrounded by a monolayer epithelium of nearly 1000 follicle cells. At stage 8 of oogenesis, these cells initiate invading the neighbouring nurse cells, and reach the oocyte boundary by Stage 10.

Krüppel is a gap gene in Drosophila melanogaster, located on the 2R chromosome, which encodes a zinc finger C2H2 transcription factor. Gap genes work together to establish the anterior-posterior segment patterning of the insect through regulation of the transcription factor encoding pair rule genes. These genes in turn regulate segment polarity genes. Krüppel means "cripple" in German, named for the crippled appearance of mutant larvae, who have failed to develop proper thoracic and anterior segments in the abdominal region. Mutants can also have abdominal mirror duplications.

A gap gene is a type of gene involved in the development of the segmented embryos of some arthropods. Gap genes are defined by the effect of a mutation in that gene, which causes the loss of contiguous body segments, resembling a gap in the normal body plan. Each gap gene, therefore, is necessary for the development of a section of the organism.
In the field of developmental biology, regional differentiation is the process by which different areas are identified in the development of the early embryo. The process by which the cells become specified differs between organisms.

The bicoid 3′-UTR regulatory element is an mRNA regulatory element that controls the gene expression of the bicoid protein in fruitfly Drosophila melanogaster.
oskar is a gene required for the development of the Drosophila embryo. It defines the posterior pole during early embryogenesis. Its two isoforms, short and long, play different roles in Drosophila embryonic development. oskar was named after the main character from the Günter Grass novel The Tin Drum, who refuses to grow up.

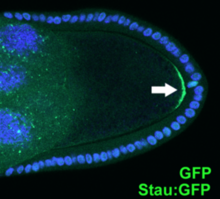
mRNA localization is a common mode of posttranscriptional regulation of gene expression that targets a protein to its site of function. Proteins are highly dependent on cellular environments for stability and function, therefore, mRNA localization signals are crucial for maintaining protein function. The Gurken localisation signal is an RNA regulatory element conserved across many species of Drosophila. The element consists of an RNA stem loop within the coding region of the messenger RNA that forms a signal for dynein-mediated Gurken mRNA transport to the dorsoanterior cap near the nucleus of the oocyte.

Double-stranded RNA-binding protein Staufen homolog 1 is a protein that in humans is encoded by the STAU1 gene.
Protein mago nashi homolog is a human protein encoded by the MAGOH gene. This gene encodes the mammalian homolog of the Drosophila mago nashi gene. In mammals, mRNA expression is not limited to the germplasm, but is ubiquitous in adult tissues and can be induced by serum stimulation of quiescent fibroblasts.

DnaJ homolog subfamily C member 3 is a protein that in humans is encoded by the DNAJC3 gene.

Centromere/kinetochore protein zw10 homolog is a protein that in humans is encoded by the ZW10 gene. This gene encodes a protein that is one of many involved in mechanisms to ensure proper chromosome segregation during cell division. The encoded protein binds to centromeres during the prophase, metaphase, and early anaphase cell division stages and to kinetochore microtubules during metaphase.

Ribosome-binding protein 1, also referred to as p180, is a protein that in humans is encoded by the RRBP1 gene.

Double-stranded RNA-binding protein Staufen homolog 2 is a protein that in humans is encoded by the STAU2 gene.
Orthodenticle (otd) is a homeobox gene found in Drosophila that regulates the development of anterior patterning, with particular involvement in the central nervous system function and eye development. It is located on the X chromosome. The gene is an ortholog of the human OTX1/OTX2 gene.
Vasa is an RNA binding protein with an ATP-dependent RNA helicase that is a member of the DEAD box family of proteins. The vasa gene is essential for germ cell development and was first identified in Drosophila melanogaster, but has since been found to be conserved in a variety of vertebrates and invertebrates including humans. The Vasa protein is found primarily in germ cells in embryos and adults, where it is involved in germ cell determination and function, as well as in multipotent stem cells, where its exact function is unknown.
The XMAP215/Dis1 family is a highly conserved group of microtubule-associated proteins (MAPs) in eukaryotic organisms. These proteins are unique MAPs because they primarily interact with the growing-end (plus-end) of microtubules. This special property classifies this protein family as plus-end tracking proteins (+TIPs).
The gene Maelstrom, Mael, creates a protein, which was first located in Drosophila melanogaster in the nuage perinuclear structure and has functionality analogous to the spindle, spn, gene class. Its mammalian homolog is MAEL.
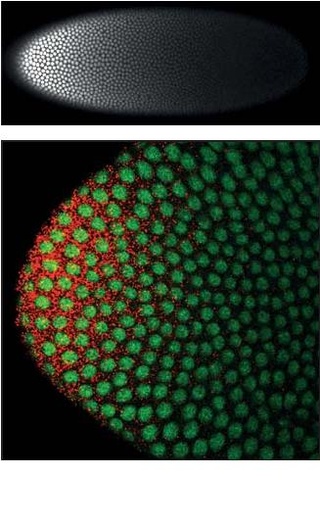
Homeotic protein bicoid is encoded by the bcd maternal effect gene in Drosophilia. Homeotic protein bicoid concentration gradient patterns the anterior-posterior (A-P) axis during Drosophila embryogenesis. Bicoid was the first protein demonstrated to act as a morphogen. Although bicoid is important for the development of Drosophila and other higher dipterans, it is absent from most other insects, where its role is accomplished by other genes.
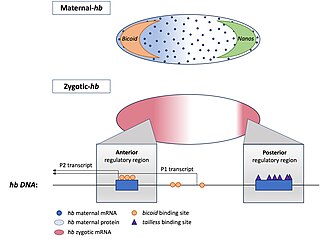
Hunchback is a maternal effect and zygotic gene expressed in the embryos of the fruit fly Drosophila melanogaster. In maternal effect genes, the RNA or protein from the mother’s gene is deposited into the oocyte or embryo before the embryo can express its own zygotic genes.