Related Research Articles

Exodeoxyribonuclease V is an enzyme of E. coli that initiates recombinational repair from potentially lethal double strand breaks in DNA which may result from ionizing radiation, replication errors, endonucleases, oxidative damage, and a host of other factors. The RecBCD enzyme is both a helicase that unwinds, or separates the strands of DNA, and a nuclease that makes single-stranded nicks in DNA. It catalyses exonucleolytic cleavage in either 5′- to 3′- or 3′- to 5′-direction to yield 5′-phosphooligonucleotides.

RecA is a 38 kilodalton protein essential for the repair and maintenance of DNA. A RecA structural and functional homolog has been found in every species in which one has been seriously sought and serves as an archetype for this class of homologous DNA repair proteins. The homologous protein is called RAD51 in eukaryotes and RadA in archaea.
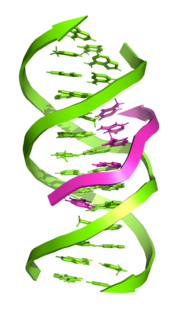
Triple-stranded DNA is a DNA structure in which three oligonucleotides wind around each other and form a triple helix. In triple-stranded DNA, the third strand binds to a B-form DNA double helix by forming Hoogsteen base pairs or reversed Hoogsteen hydrogen bonds.

Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids. It is widely used by cells to accurately repair harmful breaks that occur on both strands of DNA, known as double-strand breaks (DSB), in a process called homologous recombinational repair (HRR). Homologous recombination also produces new combinations of DNA sequences during meiosis, the process by which eukaryotes make gamete cells, like sperm and egg cells in animals. These new combinations of DNA represent genetic variation in offspring, which in turn enables populations to adapt during the course of evolution. Homologous recombination is also used in horizontal gene transfer to exchange genetic material between different strains and species of bacteria and viruses.
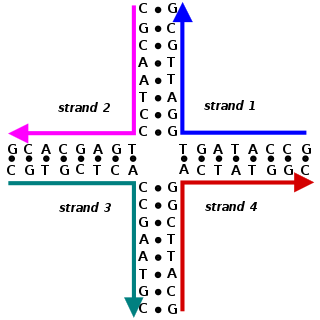
A Holliday junction is a branched nucleic acid structure that contains four double-stranded arms joined. These arms may adopt one of several conformations depending on buffer salt concentrations and the sequence of nucleobases closest to the junction. The structure is named after Robin Holliday, the molecular biologist who proposed its existence in 1964.
Recombinases are genetic recombination enzymes.

Bloom syndrome protein is a protein that in humans is encoded by the BLM gene and is not expressed in Bloom syndrome.

RAD52 homolog , also known as RAD52, is a protein which in humans is encoded by the RAD52 gene.

DNA repair and recombination protein RAD54-like is a protein that in humans is encoded by the RAD54L gene.
SAE2 is a gene in budding yeast, coding for the protein Sae2, which is involved in DNA repair. Sae2 is a part of the homologous recombination process in response to double-strand breaks. It is best characterized in the yeast model organism Saccharomyces cerevisiae. Homologous genes in other organisms include Ctp1 in fission yeast, Com1 in plants, and CtIP in higher eukaryotes including humans.
DNA topoisomerase 3-alpha is an enzyme that in humans is encoded by the TOP3A gene.

Meiotic recombination protein DMC1/LIM15 homolog is a protein that in humans is encoded by the DMC1 gene.

Crossover junction endonuclease MUS81 is an enzyme that in humans is encoded by the MUS81 gene.

DNA mismatch repair protein Mlh3 is a protein that in humans is encoded by the MLH3 gene.

RecQ-mediated genome instability protein 1 is a protein that in humans is encoded by the RMI1 gene.
Microhomology-mediated end joining (MMEJ), also known as alternative nonhomologous end-joining (Alt-NHEJ) is one of the pathways for repairing double-strand breaks in DNA. As reviewed by McVey and Lee, the foremost distinguishing property of MMEJ is the use of microhomologous sequences during the alignment of broken ends before joining, thereby resulting in deletions flanking the original break. MMEJ is frequently associated with chromosome abnormalities such as deletions, translocations, inversions and other complex rearrangements.
Sgs1, also known as slow growth suppressor 1, is a DNA helicase protein found in Saccharomyces cerevisiae. It is a homolog of the bacterial RecQ helicase. Like the other members of the RecQ helicase family, Sgs1 is important for DNA repair. In particular, Sgs1 collaborates with other proteins to repair double-strand breaks during homologous recombination in eukaryotes.
The RecF pathway, also called the RecFOR pathway, is a pathway of homologous recombination that repairs DNA in bacteria. It repairs breaks that occur on only one of DNA's two strands, known as single-strand gaps. The RecF pathway can also repair double-strand breaks in DNA when the RecBCD pathway, another pathway of homologous recombination in bacteria, is inactivated by mutations. Like the RecBCD pathway, the RecF pathway requires RecA for strand invasion. The two pathways are also similar in their phases of branch migration, in which the Holliday junction slides in one direction, and resolution, in which the Holliday junctions are cleaved apart by enzymes.
Conformational proofreading or conformational selection is a general mechanism of molecular recognition systems in which introducing a structural mismatch between a molecular recognizer and its target, or an energetic barrier, enhances the recognition specificity and quality. Conformational proofreading does not require the consumption of energy and may therefore be used in any molecular recognition system. Conformational proofreading is especially useful in scenarios where the recognizer has to select the appropriate target among many similar competitors.

Synthesis-dependent strand annealing (SDSA) is a major mechanism of homology-directed repair of DNA double-strand breaks (DSBs). Although many of the features of SDSA were first suggested in 1976, the double-Holliday junction model proposed in 1983 was favored by many researchers. In 1994, studies of double-strand gap repair in Drosophila were found to be incompatible with the double-Holliday junction model, leading researchers to propose a model they called synthesis-dependent strand annealing. Subsequent studies of meiotic recombination in S. cerevisiae found that non-crossover products appear earlier than double-Holliday junctions or crossover products, challenging the previous notion that both crossover and non-crossover products are produced by double-Holliday junctions and leading the authors to propose that non-crossover products are generated through SDSA.
References
- ↑ "UC Davis College of Biological Sciences" . Retrieved 22 February 2013.
- ↑ http://news.ucdavis.edu/search/news_detail.lasso?id=8149 . Retrieved 22 February 2013.
{{cite news}}: Missing or empty|title=(help) - ↑ http://chemistry.georgetown.edu/news/news_11.06.07.html . Retrieved 22 February 2013.
{{cite news}}: Missing or empty|title=(help) - ↑ Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB (November 11, 2004). "Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks". Nature. 432 (7014): 187–93. Bibcode:2004Natur.432..187S. doi:10.1038/nature02988. PMID 15538360. S2CID 2916995.
- ↑ Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC (September 2, 2010). "DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2". Nature. 467 (7311): 112–6. Bibcode:2010Natur.467..112C. doi:10.1038/nature09355. PMC 3089589 . PMID 20811461.
- ↑ Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC (October 2019). "Direct observation of individual RecA filaments assembling on single DNA molecules". Nature. 443 (7113): 875–8. Bibcode:2006Natur.443..875G. doi:10.1038/nature05197. PMID 16988658. S2CID 4416295.
- ↑ Bell JC, Plank JL, Dombrowski CC, Kowalczykowski SC (November 8, 2012). "Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA". Nature. 491 (7423): 274–8. Bibcode:2012Natur.491..274B. doi:10.1038/nature11598. PMC 4112059 . PMID 23103864.
- ↑ Jensen RB, Carreira A, Kowalczykowski SC (October 7, 2010). "Purified human BRCA2 stimulates RAD51-mediated recombination". Nature. 467 (7316): 678–83. Bibcode:2010Natur.467..678J. doi:10.1038/nature09399. PMC 2952063 . PMID 20729832.
- ↑ "Nature News" . Retrieved 22 February 2013.
- ↑ Cejka P, Plank JL, Bachrati CZ, Hickson ID, Kowalczykowski SC (Nov 2010). "Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3". Nat Struct Mol Biol. 17 (11): 1377–82. doi:10.1038/nsmb.1919. PMC 2988882 . PMID 20935631.
- ↑ Cejka P, Plank JL, Dombrowski CC, Kowalczykowski SC (September 28, 2012). "Decatenation of DNA by the S. cerevisiae Sgs1-Top3-Rmi1 and RPA complex: a mechanism for disentangling chromosomes". Molecular Cell. 47 (6): 886–96. doi:10.1016/j.molcel.2012.06.032. PMC 3462259 . PMID 22885009.
- ↑ Forget AL, Kowalczykowski SC (February 8, 2012). "Single-molecule imaging of DNA pairing by RecA reveals a three-dimensional homology search". Nature. 482 (7385): 423–7. Bibcode:2012Natur.482..423F. doi:10.1038/nature10782. PMC 3288143 . PMID 22318518.