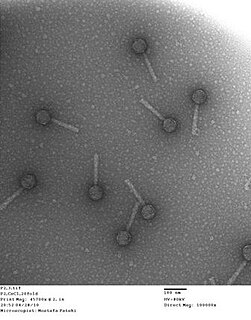
A bacteriophage, also known informally as a phage, is a virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν, meaning "to devour". Bacteriophages are composed of proteins that encapsulate a DNA or RNA genome, and may have structures that are either simple or elaborate. Their genomes may encode as few as four genes and as many as hundreds of genes. Phages replicate within the bacterium following the injection of their genome into its cytoplasm.
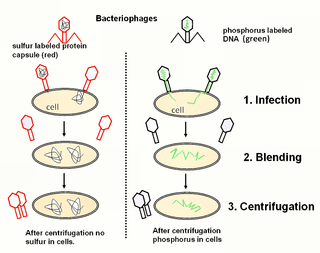
The Hershey–Chase experiments were a series of experiments conducted in 1952 by Alfred Hershey and Martha Chase that helped to confirm that DNA is genetic material. While DNA had been known to biologists since 1869, many scientists still assumed at the time that proteins carried the information for inheritance because DNA appeared to be an inert molecule, and, since it is located in the nucleus, its role was considered to be phosphorus storage. In their experiments, Hershey and Chase showed that when bacteriophages, which are composed of DNA and protein, infect bacteria, their DNA enters the host bacterial cell, but most of their protein does not. Hershey and Chase and subsequent discoveries all served to prove that DNA is the hereditary material.

Virology is the study of viral – submicroscopic, parasitic particles of genetic material contained in a protein coat – and virus-like agents. It focuses on the following aspects of viruses: their structure, classification and evolution, their ways to infect and exploit host cells for reproduction, their interaction with host organism physiology and immunity, the diseases they cause, the techniques to isolate and culture them, and their use in research and therapy. Virology is considered to be a subfield of microbiology or of medicine.
Virulence is a pathogen's or microbe's ability to infect or damage a host.

Transduction is the process by which foreign DNA is introduced into an eukaryotic cell by a virus or viral vector. An example is the viral transfer of DNA from one bacterium to another and hence an example of horizontal gene transfer. Transduction does not require physical contact between the cell donating the DNA and the cell receiving the DNA, and it is DNase resistant. Transduction is a common tool used by molecular biologists to stably introduce a foreign gene into a host cell's genome.

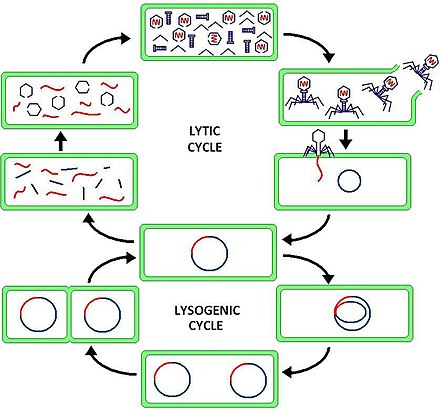
The lytic cycle is one of the two cycles of viral reproduction, the other being the lysogenic cycle. The lytic cycle results in the destruction of the infected cell and its membrane. Bacteriophages that only use the lytic cycle are called virulent phages.
Escherichia virus T4 is a species of bacteriophages that infect Escherichia coli bacteria. It is a member of virus subfamily Tevenvirinae and includes among other strains Enterobacteria phage T2, Enterobacteria phage T4 and Enterobacteria phage T6. T4 is capable of undergoing only a lytic lifecycle and not the lysogenic lifecycle.
Microviridae is a family of bacteriophages with a single-stranded DNA genome. The name of this family is derived from the ancient Greek word μικρός (mikrós), meaning "small". This refers to the size of their genomes, which are among the smallest of the DNA viruses. Enterobacteria, intracellular parasitic bacteria, and spiroplasma serve as natural hosts. There are currently 12 species in this family, divided among 7 genera and one subfamily.

Molecular virology is the study of viruses on a molecular level. Viruses are submicroscopic parasites that replicate inside host cells. They are able to successfully infect and parasitize all kinds of life forms- from microorganisms to plants and animals- and as a result viruses have more biological diversity than the rest of the bacterial, plant, and animal kingdoms combined. Studying this diversity is the key to a better understanding of how viruses interact with their hosts, replicate inside them, and cause diseases.

The Caudovirales are an order of viruses also known as the tailed bacteriophages. Under the Baltimore classification scheme, the Caudovirales are group I viruses as they have double stranded DNA (dsDNA) genomes, which can be anywhere from 18,000 base pairs to 500,000 base pairs in length. The virus particles have a distinct shape; each virion has an icosahedral head that contains the viral genome, and is attached to a flexible tail by a connector protein. The order encompasses a wide range of viruses, many containing genes of similar nucleotide sequence and function. However, some tailed bacteriophage genomes can vary quite significantly in nucleotide sequence, even among the same genus. Due to their characteristic structure and possession of potentially homologous genes, it is believed these bacteriophages possess a common origin.

Lysogeny, or the lysogenic cycle, is one of two cycles of viral reproduction. Lysogeny is characterized by integration of the bacteriophage nucleic acid into the host bacterium's genome or formation of a circular replicon in the bacterial cytoplasm. In this condition the bacterium continues to live and reproduce normally. The genetic material of the bacteriophage, called a prophage, can be transmitted to daughter cells at each subsequent cell division, and at later events can release it, causing proliferation of new phages via the lytic cycle. Lysogenic cycles can also occur in eukaryotes, although the method of DNA incorporation is not fully understood.
Bacteriophage T7 is a bacteriophage, a virus composed of DNA. It infects most strains of an Escherichia coli and relies on these hosts to propagate. Bacteriophage T7 has a lytic life cycle, meaning that it destroys the cell it infects. It also possesses several properties that make it an ideal phage for experimentation: its purification and concentration have produced consistent values in chemical analyses; it can be rendered noninfectious by exposure to UV light; and it can be used in phage display to clone RNA binding proteins.

Viral entry is the earliest stage of infection in the viral life cycle, as the virus comes into contact with the host cell and introduces viral material into the cell. The major steps involved in viral entry are shown below. Despite the variation among viruses, there are several shared generalities concerning viral entry.
Enterobacteria phage P22 is a bacteriophage in the Podoviridae family that infects Salmonella typhimurium. Like many phages, it has been used in molecular biology to induce mutations in cultured bacteria and to introduce foreign genetic material. P22 has been used in generalized transduction and is an important tool for investigating Salmonella genetics.

Tectiviridae is a family of viruses with three genera. Gram-negative bacteria serve as natural hosts. There are currently four species in this genus including the type species Enterobacteria phage PRD1. Tectiviruses have no head-tail structure, but are capable of producing tail-like tubes of ~ 60×10 nm upon adsorption or after chloroform treatment. The name is derived from Latin tectus.

Corticovirus is a genus of viruses in the family Corticoviridae. Corticoviruses are bacteriophages; that is, their natural hosts are bacteria. The genus contains only one species, the type species Pseudoalteromonas virus PM2. The name is derived from Latin cortex, corticis. However, prophages closely related to PM2 are abundant in the genomes of aquatic bacteria, suggesting that the ecological importance of corticoviruses might be underestimated. Bacteriophage PM2 was first described in 1968 after isolation from seawater sampled from the coast of Chile.

A virus is a submicroscopic infectious agent that replicates only inside the living cells of an organism. Viruses can infect all types of life forms, from animals and plants to microorganisms, including bacteria and archaea. Since Dmitri Ivanovsky's 1892 article describing a non-bacterial pathogen infecting tobacco plants, and the discovery of the tobacco mosaic virus by Martinus Beijerinck in 1898, about 5,000 virus species have been described in detail, of the millions of types of viruses in the environment. Viruses are found in almost every ecosystem on Earth and are the most numerous type of biological entity. The study of viruses is known as virology, a subspeciality of microbiology.
Bacteriophage P2, scientific name Escherichia virus P2, is a temperate phage that infects E. coli. It is a tailed virus with a contractile sheath and is thus classified in the genus P2virus, subfamily Peduovirinae, family Myoviridae within order Caudovirales. This genus of viruses includes many P2-like phages as well as the satellite phage P4.

Spiroplasma phage 1-R8A2B is a filamentous bacteriophage in the genus Plectrovirus of the family Inoviridae, part of the group of single-stranded DNA viruses. The virus has many synonyms, such as SpV1-R8A2 B, Spiroplasma phage 1, and Spiroplasma virus 1, SpV1. SpV1-R8A2 B infects Spiroplasma citri. Its host itself is a prokaryotic pathogen for citrus plants, causing Citrus stubborn disease.
Escherichia virus CC31, formerly known as Enterobacter virus CC31, is a dsDNA bacteriophage of the subfamily Tevenvirinae responsible for infecting the bacteria family of Enterobacteriaceae. It is one of two discovered viruses of the genus Karamvirus, diverging away from the previously discovered T4virus, as a clonal complex (CC). CC31 was first isolated from Escherichia coli B strain S/6/4 and is primarily associated with Escherichia, even though is named after Enterobacter.