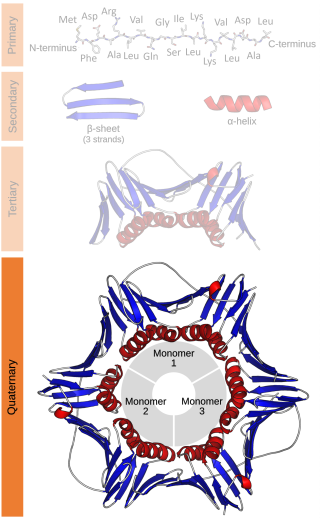
Protein quaternary structure is the fourth classification level of protein structure. Protein quaternary structure refers to the structure of proteins which are themselves composed of two or more smaller protein chains. Protein quaternary structure describes the number and arrangement of multiple folded protein subunits in a multi-subunit complex. It includes organizations from simple dimers to large homooligomers and complexes with defined or variable numbers of subunits. In contrast to the first three levels of protein structure, not all proteins will have a quaternary structure since some proteins function as single units. Protein quaternary structure can also refer to biomolecular complexes of proteins with nucleic acids and other cofactors.

The major histocompatibility complex (MHC) is a large locus on vertebrate DNA containing a set of closely linked polymorphic genes that code for cell surface proteins essential for the adaptive immune system. These cell surface proteins are called MHC molecules.

A protein complex or multiprotein complex is a group of two or more associated polypeptide chains. Protein complexes are distinct from multidomain enzymes, in which multiple catalytic domains are found in a single polypeptide chain.
In biochemistry, biotinylation is the process of covalently attaching biotin to a protein, nucleic acid or other molecule. Biotinylation is rapid, specific and is unlikely to disturb the natural function of the molecule due to the small size of biotin. Biotin binds to streptavidin and avidin with an extremely high affinity, fast on-rate, and high specificity, and these interactions are exploited in many areas of biotechnology to isolate biotinylated molecules of interest. Biotin-binding to streptavidin and avidin is resistant to extremes of heat, pH and proteolysis, making capture of biotinylated molecules possible in a wide variety of environments. Also, multiple biotin molecules can be conjugated to a protein of interest, which allows binding of multiple streptavidin, avidin or neutravidin protein molecules and increases the sensitivity of detection of the protein of interest. There is a large number of biotinylation reagents available that exploit the wide range of possible labelling methods. Due to the strong affinity between biotin and streptavidin, the purification of biotinylated proteins has been a widely used approach to identify protein-protein interactions and post-translational events such as ubiquitylation in molecular biology.
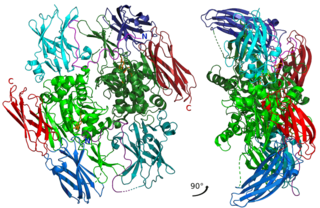
Factor XIII or fibrin stabilizing factor is a zymogen found in blood of humans and some other animals. It is activated by thrombin to factor XIIIa. Factor XIIIa is an enzyme of the blood coagulation system that crosslinks fibrin. Deficiency of XIII worsens clot stability and increases bleeding tendency.

Streptavidin is a 52 kDa protein (tetramer) purified from the bacterium Streptomyces avidinii. Streptavidin homo-tetramers have an extraordinarily high affinity for biotin. With a dissociation constant (Kd) on the order of ≈10−14 mol/L, the binding of biotin to streptavidin is one of the strongest non-covalent interactions known in nature. Streptavidin is used extensively in molecular biology and bionanotechnology due to the streptavidin-biotin complex's resistance to organic solvents, denaturants, detergents, proteolytic enzymes, and extremes of temperature and pH.

MHC class I molecules are one of two primary classes of major histocompatibility complex (MHC) molecules and are found on the cell surface of all nucleated cells in the bodies of vertebrates. They also occur on platelets, but not on red blood cells. Their function is to display peptide fragments of proteins from within the cell to cytotoxic T cells; this will trigger an immediate response from the immune system against a particular non-self antigen displayed with the help of an MHC class I protein. Because MHC class I molecules present peptides derived from cytosolic proteins, the pathway of MHC class I presentation is often called cytosolic or endogenous pathway.
A dodecameric protein has a quaternary structure consisting of 12 protein subunits in a complex. Dodecameric complexes can have a number of subunit 'topologies', but typically only a few of the theoretically possible subunit arrangements are observed in protein structures.

Argininosuccinate synthase or synthetase is an enzyme that catalyzes the synthesis of argininosuccinate from citrulline and aspartate. In humans, argininosuccinate synthase is encoded by the ASS gene located on chromosome 9.

The enzyme argininosuccinate lyase (EC 4.3.2.1, ASL, argininosuccinase; systematic name 2-(N ω-L-arginino)succinate arginine-lyase (fumarate-forming)) catalyzes the reversible breakdown of argininosuccinate:
Acyl-CoA dehydrogenases (ACADs) are a class of enzymes that function to catalyze the initial step in each cycle of fatty acid β-oxidation in the mitochondria of cells. Their action results in the introduction of a trans double-bond between C2 (α) and C3 (β) of the acyl-CoA thioester substrate. Flavin adenine dinucleotide (FAD) is a required co-factor in addition to the presence of an active site glutamate in order for the enzyme to function.

Antigen presentation is a vital immune process that is essential for T cell immune response triggering. Because T cells recognize only fragmented antigens displayed on cell surfaces, antigen processing must occur before the antigen fragment can be recognized by a T-cell receptor. Specifically, the fragment, bound to the major histocompatibility complex (MHC), is transported to the surface of the cell, a process known as presentation. If there has been an infection with viruses or bacteria, the cell will present an endogenous or exogenous peptide fragment derived from the antigen by MHC molecules. There are two types of MHC molecules which differ in the behaviour of the antigens: MHC class I molecules (MHC-I) bind peptides from the cell cytosol, while peptides generated in the endocytic vesicles after internalisation are bound to MHC class II (MHC-II). Cellular membranes separate these two cellular environments - intracellular and extracellular. Each T cell can only recognize tens to hundreds of copies of a unique sequence of a single peptide among thousands of other peptides presented on the same cell, because an MHC molecule in one cell can bind to quite a large range of peptides. Predicting which antigens will be presented to the immune system by a certain MHC/HLA type is difficult, but the technology involved is improving.

Propionyl-CoA carboxylase (EC 6.4.1.3, PCC) catalyses the carboxylation reaction of propionyl-CoA in the mitochondrial matrix. PCC has been classified both as a ligase and a lyase. The enzyme is biotin-dependent. The product of the reaction is (S)-methylmalonyl CoA.

Sorbitol dehydrogenase is a cytosolic enzyme. In humans this protein is encoded by the SORD gene.

Minor histocompatibility antigen are peptides presented on the cellular surface of donated organs that are known to give an immunological response in some organ transplants. They cause problems of rejection less frequently than those of the major histocompatibility complex (MHC). Minor histocompatibility antigens (MiHAs) are diverse, short segments of proteins and are referred to as peptides. These peptides are normally around 9-12 amino acids in length and are bound to both the major histocompatibility complex (MHC) class I and class II proteins. Peptide sequences can differ among individuals and these differences arise from SNPs in the coding region of genes, gene deletions, frameshift mutations, or insertions. About a third of the characterized MiHAs come from the Y chromosome. Prior to becoming a short peptide sequence, the proteins expressed by these polymorphic or diverse genes need to be digested in the proteasome into shorter peptides. These endogenous or self peptides are then transported into the endoplasmic reticulum with a peptide transporter pump called TAP where they encounter and bind to the MHC class I molecule. This contrasts with MHC class II molecules's antigens which are peptides derived from phagocytosis/endocytosis and molecular degradation of non-self entities' proteins, usually by antigen-presenting cells. MiHA antigens are either ubiquitously expressed in most tissue like skin and intestines or restrictively expressed in the immune cells.
A tetramer assay is a procedure that uses tetrameric proteins to detect and quantify T cells that are specific for a given antigen within a blood sample. The tetramers used in the assay are made up of four major histocompatibility complex (MHC) molecules, which are found on the surface of most cells in the body. MHC molecules present peptides to T-cells as a way to communicate the presence of viruses, bacteria, cancerous mutations, or other antigens in a cell. If a T-cell's receptor matches the peptide being presented by an MHC molecule, an immune response is triggered. Thus, MHC tetramers that are bioengineered to present a specific peptide can be used to find T-cells with receptors that match that peptide. The tetramers are labeled with a fluorophore, allowing tetramer-bound T-cells to be analyzed with flow cytometry. Quantification and sorting of T-cells by flow cytometry enables researchers to investigate immune response to viral infection and vaccine administration as well as functionality of antigen-specific T-cells. Generally, if a person's immune system has encountered a pathogen, the individual will possess T cells with specificity toward some peptide on that pathogen. Hence, if a tetramer stain specific for a pathogenic peptide results in a positive signal, this may indicate that the person's immune system has encountered and built a response to that pathogen.

Adenylosuccinate lyase is an enzyme that in humans is encoded by the ADSL gene.
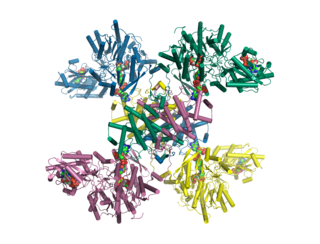
ATP citrate synthase (also ATP citrate lyase (ACLY)) is an enzyme that in animals represents an important step in fatty acid biosynthesis. By converting citrate to acetyl-CoA, the enzyme links carbohydrate metabolism, which yields citrate as an intermediate, with fatty acid biosynthesis, which consumes acetyl-CoA. In plants, ATP citrate lyase generates cytosolic acetyl-CoA precursors of thousands of specialized metabolites, including waxes, sterols, and polyketides.
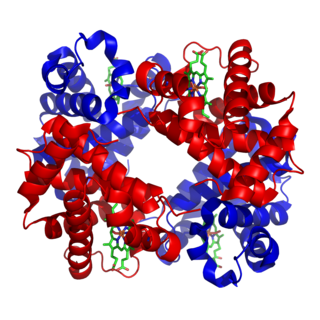
A tetramer (tetra-, "four" + -mer, "parts") is an oligomer formed from four monomers or subunits. The associated property is called tetramery. An example from inorganic chemistry is titanium methoxide with the empirical formula Ti(OCH3)4, which is tetrameric in solid state and has the molecular formula Ti4(OCH3)16. An example from organic chemistry is kobophenol A, a substance that is formed by combining four molecules of resveratrol.
MHC multimers are oligomeric forms of MHC molecules, designed to identify and isolate T-cells with high affinity to specific antigens amid a large group of unrelated T-cells. Multimers generally range in size from dimers to octamers; however, some companies use even higher quantities of MHC per multimer. Multimers may be used to display class 1 MHC, class 2 MHC, or nonclassical molecules from species such as monkeys, mice, and humans.

















