
The CpG sites or CG sites are regions of DNA where a cytosine nucleotide is followed by a guanine nucleotide in the linear sequence of bases along its 5' → 3' direction. CpG sites occur with high frequency in genomic regions called CpG islands. Cytosines in CpG dinucleotides can be methylated to form 5-methylcytosines. Enzymes that add a methyl group are called DNA methyltransferases. In mammals, 70% to 80% of CpG cytosines are methylated. Methylating the cytosine within a gene can change its expression, a mechanism that is part of a larger field of science studying gene regulation that is called epigenetics.
DNA glycosylases are a family of enzymes involved in base excision repair, classified under EC number EC 3.2.2. Base excision repair is the mechanism by which damaged bases in DNA are removed and replaced. DNA glycosylases catalyze the first step of this process. They remove the damaged nitrogenous base while leaving the sugar-phosphate backbone intact, creating an apurinic/apyrimidinic site, commonly referred to as an AP site. This is accomplished by flipping the damaged base out of the double helix followed by cleavage of the N-glycosidic bond.

Oxidative stress reflects an imbalance between the systemic manifestation of reactive oxygen species and a biological system's ability to readily detoxify the reactive intermediates or to repair the resulting damage. Disturbances in the normal redox state of cells can cause toxic effects through the production of peroxides and free radicals that damage all components of the cell, including proteins, lipids, and DNA. Oxidative stress from oxidative metabolism causes base damage, as well as strand breaks in DNA. Base damage is mostly indirect and caused by reactive oxygen species (ROS) generated, e.g. O2− (superoxide radical), OH (hydroxyl radical) and H2O2 (hydrogen peroxide). Further, some reactive oxidative species act as cellular messengers in redox signaling. Thus, oxidative stress can cause disruptions in normal mechanisms of cellular signaling.

Base excision repair (BER) is a cellular mechanism, studied in the fields of biochemistry and genetics, that repairs damaged DNA throughout the cell cycle. It is responsible primarily for removing small, non-helix-distorting base lesions from the genome. The related nucleotide excision repair pathway repairs bulky helix-distorting lesions. BER is important for removing damaged bases that could otherwise cause mutations by mispairing or lead to breaks in DNA during replication. BER is initiated by DNA glycosylases, which recognize and remove specific damaged or inappropriate bases, forming AP sites. These are then cleaved by an AP endonuclease. The resulting single-strand break can then be processed by either short-patch or long-patch BER.
DNA oxidation is the process of oxidative damage of deoxyribonucleic acid. As described in detail by Burrows et al., 8-oxo-2'-deoxyguanosine (8-oxo-dG) is the most common oxidative lesion observed in duplex DNA because guanine has a lower one-electron reduction potential than the other nucleosides in DNA. The one electron reduction potentials of the nucleosides are guanine 1.29, adenine 1.42, cytosine 1.6 and thymine 1.7. About 1 in 40,000 guanines in the genome are present as 8-oxo-dG under normal conditions. This means that >30,000 8-oxo-dGs may exist at any given time in the genome of a human cell. Another product of DNA oxidation is 8-oxo-dA. 8-oxo-dA occurs at about 1/3 the frequency of 8-oxo-dG. The reduction potential of guanine may be reduced by as much as 50%, depending on the particular neighboring nucleosides stacked next to it within DNA.
Cell damage is a variety of changes of stress that a cell suffers due to external as well as internal environmental changes. Amongst other causes, this can be due to physical, chemical, infectious, biological, nutritional or immunological factors. Cell damage can be reversible or irreversible. Depending on the extent of injury, the cellular response may be adaptive and where possible, homeostasis is restored. Cell death occurs when the severity of the injury exceeds the cell's ability to repair itself. Cell death is relative to both the length of exposure to a harmful stimulus and the severity of the damage caused. Cell death may occur by necrosis or apoptosis.
A DNA repair-deficiency disorder is a medical condition due to reduced functionality of DNA repair.

In genetics, crosslinking of DNA occurs when various exogenous or endogenous agents react with two nucleotides of DNA, forming a covalent linkage between them. This crosslink can occur within the same strand (intrastrand) or between opposite strands of double-stranded DNA (interstrand). These adducts interfere with cellular metabolism, such as DNA replication and transcription, triggering cell death. These crosslinks can, however, be repaired through excision or recombination pathways.
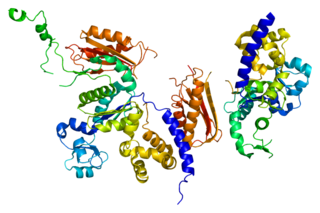
DNA polymerase kappa is an DNA polymerase that in humans is encoded by the POLK gene. It is involved in translesion synthesis.

G/T mismatch-specific thymine DNA glycosylase is an enzyme that in humans is encoded by the TDG gene. Several bacterial proteins have strong sequence homology with this protein.

Endonuclease III-like protein 1 is an enzyme that in humans is encoded by the NTHL1 gene.
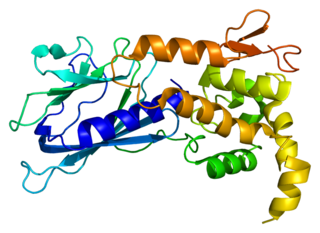
Endonuclease VIII-like 1 is an enzyme that in humans is encoded by the NEIL1 gene.

Peptide methionine sulfoxide reductase (Msr) is a family of enzymes that in humans is encoded by the MSRA gene.

DNA polymerase eta, is a protein that in humans is encoded by the POLH gene.
The DNA damage theory of aging proposes that aging is a consequence of unrepaired accumulation of naturally occurring DNA damages. Damage in this context is a DNA alteration that has an abnormal structure. Although both mitochondrial and nuclear DNA damage can contribute to aging, nuclear DNA is the main subject of this analysis. Nuclear DNA damage can contribute to aging either indirectly or directly.

8-Oxo-2'-deoxyguanosine (8-oxo-dG) is an oxidized derivative of deoxyguanosine. 8-Oxo-dG is one of the major products of DNA oxidation. Concentrations of 8-oxo-dG within a cell are a measurement of oxidative stress.

The FPG IleRS zinc finger domain represents a zinc finger domain found at the C-terminal in both DNA glycosylase/AP lyase enzymes and in isoleucyl tRNA synthetase. In these two types of enzymes, the C-terminal domain forms a zinc finger.
Genome instability refers to a high frequency of mutations within the genome of a cellular lineage. These mutations can include changes in nucleic acid sequences, chromosomal rearrangements or aneuploidy. Genome instability does occur in bacteria. In multicellular organisms genome instability is central to carcinogenesis, and in humans it is also a factor in some neurodegenerative diseases such as amyotrophic lateral sclerosis or the neuromuscular disease myotonic dystrophy.

In molecular biology, the H2TH domain is a DNA-binding domain found in DNA glycosylase/AP lyase enzymes, which are involved in base excision repair of DNA damaged by oxidation or by mutagenic agents. Most damage to bases in DNA is repaired by the base excision repair pathway. These enzymes are primarily from bacteria, and have both DNA glycosylase activity EC 3.2.2.- and AP lyase activity EC 4.2.99.18. Examples include formamidopyrimidine-DNA glycosylases and endonuclease VIII (Nei).
DNA damage is distinctly different from mutation, although both are types of error in DNA. DNA damage is an abnormal chemical structure in DNA, while a mutation is a change in the sequence of standard base pairs. DNA damages cause changes in the structure of the genetic material and prevents the replication mechanism from functioning and performing properly.












