Related Research Articles

Metagenomics is the study of genetic material recovered directly from environmental samples. The broad field may also be referred to as environmental genomics, ecogenomics or community genomics.

The International Committee on Taxonomy of Viruses (ICTV) authorizes and organizes the taxonomic classification of and the nomenclatures for viruses. The ICTV has developed a universal taxonomic scheme for viruses, and thus has the means to appropriately describe, name, and classify every virus that affects living organisms. The members of the International Committee on Taxonomy of Viruses are considered expert virologists. The ICTV was formed from and is governed by the Virology Division of the International Union of Microbiological Societies. Detailed work, such as delimiting the boundaries of species within a family, typically is performed by study groups of experts in the families.
Anelloviridae is a family of viruses. They are classified as vertebrate viruses and have a non-enveloped capsid, which is round with isometric, icosahedral symmetry and has a triangulation number of 3.

A virus is a submicroscopic infectious agent that replicates only inside the living cells of an organism. Viruses infect all life forms, from animals and plants to microorganisms, including bacteria and archaea. Since Dmitri Ivanovsky's 1892 article describing a non-bacterial pathogen infecting tobacco plants and the discovery of the tobacco mosaic virus by Martinus Beijerinck in 1898, more than 9,000 virus species have been described in detail of the millions of types of viruses in the environment. Viruses are found in almost every ecosystem on Earth and are the most numerous type of biological entity. The study of viruses is known as virology, a subspeciality of microbiology.

Forest Rohwer is an American microbial ecologist and Professor of Biology at San Diego State University. His particular interests include coral reef microbial ecology and viruses as both evolutionary agents and opportunistic pathogens in various environments.
Biological dark matter is an informal term for unclassified or poorly understood genetic material. This genetic material may refer to genetic material produced by unclassified microorganisms. By extension, biological dark matter may also refer to the un-isolated microorganism whose existence can only be inferred from the genetic material that they produce. Some of the genetic material may not fall under the three existing domains of life: Bacteria, Archaea and Eukaryota; thus, it has been suggested that a possible fourth domain of life may yet to be discovered, although other explanations are also probable. Alternatively, the genetic material may refer to non-coding DNA and non-coding RNA produced by known organisms.

The human virome is the total collection of viruses in and on the human body. Viruses in the human body may infect both human cells and other microbes such as bacteria. Some viruses cause disease, while others may be asymptomatic. Certain viruses are also integrated into the human genome as proviruses or endogenous viral elements.

Viral metagenomics is the study of viral genetic material sourced directly from the environment rather than from a host or natural reservoir. The goal is to ascertain the viral diversity in the environment that is often missed in studies targeting specific potential reservoirs. It reveals important information on virus evolution and the genetic diversity of the viral community without the need for isolating viral species and cultivating them in the laboratory. With the new techniques available that exploit next-generation sequencing (NGS), it is possible to study the virome of some ecosystems, even if the analysis still has some issues, in particular the lack of universal markers. Some of the first metagenomic studies of viruses were done with ocean samples, and revealed that most of the sequences of DNA and RNA viruses had no matches in databases. Subsequently, some studies about the soil virome were performed with a particular interest on bacteriophages, and it was discovered that there are almost the same number of viruses and bacteria. This approach has created improvements in molecular epidemiology and accelerated the discovery of novel viruses.

Virome refers to the assemblage of viruses that is often investigated and described by metagenomic sequencing of viral nucleic acids that are found associated with a particular ecosystem, organism or holobiont. The word is frequently used to describe environmental viral shotgun metagenomes. Viruses, including bacteriophages, are found in all environments, and studies of the virome have provided insights into nutrient cycling, development of immunity, and a major source of genes through lysogenic conversion.
Jingmenvirus is a group of positive-sense single-stranded RNA viruses with segmented genomes. They are primarily associated with arthropods and are the only known segmented RNA viruses that infect animal hosts. The first member of the group, the Jingmen tick virus (JMTV), was described in 2014. Another member, the Guaico Culex virus (GCXV), has a highly unusual multicomponent architecture in which the genome segments are separately enclosed in different viral capsids.
Nikos Kyrpides is a Greek-American bioscientist who has worked on the origins of life, information processing, bioinformatics, microbiology, metagenomics and microbiome data science. He is a senior staff scientist at the Berkeley National Laboratory, head of the Prokaryote Super Program and leads the Microbiome Data Science program at the US Department of Energy Joint Genome Institute.

Mya Breitbart is an American biologist and professor of biological oceanography at the University of South Florida's College of Marine Science. She is best known for her contributions to the field of viral metagenomics. Popular Science recognized her because of her approach of not trying to sequence individual viruses or organisms but to sequence everything in a given ecosystem.

Riboviria is a realm of viruses that includes all viruses that use a homologous RNA-dependent polymerase for replication. It includes RNA viruses that encode an RNA-dependent RNA polymerase, as well as reverse-transcribing viruses that encode an RNA-dependent DNA polymerase. RNA-dependent RNA polymerase (RdRp), also called RNA replicase, produces RNA from RNA. RNA-dependent DNA polymerase (RdDp), also called reverse transcriptase (RT), produces DNA from RNA. These enzymes are essential for replicating the viral genome and transcribing viral genes into messenger RNA (mRNA) for translation of viral proteins.
Clinical metagenomic next-generation sequencing (mNGS) is the comprehensive analysis of microbial and host genetic material in clinical samples from patients. It uses the techniques of metagenomics to identify and characterize the genome of bacteria, fungi, parasites, and viruses without the need for a prior knowledge of a specific pathogen directly from clinical specimens. The capacity to detect all the potential pathogens in a sample makes metagenomic next generation sequencing a potent tool in the diagnosis of infectious disease especially when other more directed assays, such as PCR, fail. Its limitations include clinical utility, laboratory validity, sense and sensitivity, cost and regulatory considerations.

Redondoviruses are a family of human-associated DNA viruses. Their name derives from the inferred circular structure of the viral genome . Redondoviruses have been identified in DNA sequence based surveys of samples from humans, primarily samples from the oral cavity and upper airway.
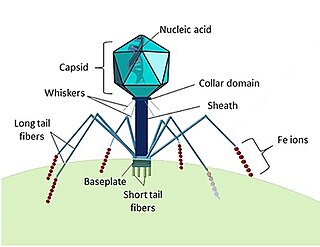
Marine viruses are defined by their habitat as viruses that are found in marine environments, that is, in the saltwater of seas or oceans or the brackish water of coastal estuaries. Viruses are small infectious agents that can only replicate inside the living cells of a host organism, because they need the replication machinery of the host to do so. They can infect all types of life forms, from animals and plants to microorganisms, including bacteria and archaea.
Lenarviricota is a phylum of RNA viruses that includes all positive-strand RNA viruses that infect prokaryotes. Some members also infect eukaryotes. Most of these viruses do not have capsids, except for the genus Ourmiavirus. The name of the group is a syllabic abbreviation of the names of founding member families "Leviviridae and Narnaviridae" with the suffix -viricota, denoting a virus phylum.

Pisuviricota is a phylum of RNA viruses that includes all positive-strand and double-stranded RNA viruses that infect eukaryotes and are not members of the phylum Kitrinoviricota,Lenarviricota or Duplornaviricota. The name of the group is a syllabic abbreviation of “picornavirus supergroup” with the suffix -viricota, indicating a virus phylum. Phylogenetic analyses suggest that Birnaviridae and Permutotetraviridae, both currently unassigned to a phylum in Orthornavirae, also belong to this phylum and that both are sister groups.
Diversity-generating retroelements (DGRs) are a family of retroelements that were first found in Bordetella phage (BPP-1), and since been found in bacteria, Archaea, Archaean viruses, temperate phages, and lytic phages. DGRs benefit their host by mutating particular regions of specific target proteins, for instance, phage tail fiber in BPP-1, lipoprotein in legionella pneumophila, and TvpA in Treponema denticola . An error-prone reverse transcriptase is responsible for generating these hypervariable regions in target proteins. In mutagenic retrohoming, a mutagenized cDNA is reverse transcribed from a template region (TR), and is replaced with a segment similar to the template region called variable region (VR). Accessory variability determinant (Avd) protein is another component of DGRs, and its complex formation with the error-prone RT is of importance to mutagenic rehoming.
Virosphere is viral part of biosphere, namely the pool of viruses in all hosts and all environments on planet earth.