Related Research Articles
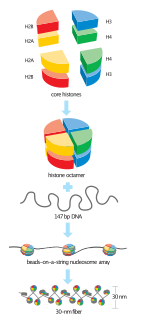
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone proteins and resembles thread wrapped around a spool. The nucleosome is the fundamental subunit of chromatin. Each nucleosome is composed of a little less than two turns of DNA wrapped around a set of eight proteins called histones, which are known as a histone octamer. Each histone octamer is composed of two copies each of the histone proteins H2A, H2B, H3, and H4.

Histone acetyltransferases (HATs) are enzymes that acetylate conserved lysine amino acids on histone proteins by transferring an acetyl group from acetyl-CoA to form ε-N-acetyllysine. DNA is wrapped around histones, and, by transferring an acetyl group to the histones, genes can be turned on and off. In general, histone acetylation increases gene expression.

In molecular biology, SWI/SNF, is a subfamily of ATP-dependent chromatin remodeling complexes, which is found in eukaryotes. In other words, it is a group of proteins that associate to remodel the way DNA is packaged. This complex is composed of several proteins – products of the SWI and SNF genes, as well as other polypeptides. It possesses a DNA-stimulated ATPase activity that can destabilize histone-DNA interactions in reconstituted nucleosomes in an ATP-dependent manner, though the exact nature of this structural change is unknown. The SWI/SNF subfamily provides crucial nucleosome rearrangement, which is seen as ejection and/or sliding. The movement of nucleosomes provides easier access to the chromatin, allowing genes to be activated or repressed.
In molecular biology and genetics, transcription coregulators are proteins that interact with transcription factors to either activate or repress the transcription of specific genes. Transcription coregulators that activate gene transcription are referred to as coactivators while those that repress are known as corepressors. The mechanism of action of transcription coregulators is to modify chromatin structure and thereby make the associated DNA more or less accessible to transcription. In humans several dozen to several hundred coregulators are known, depending on the level of confidence with which the characterisation of a protein as a coregulator can be made. One class of transcription coregulators modifies chromatin structure through covalent modification of histones. A second ATP dependent class modifies the conformation of chromatin.

Cyclic adenosine monophosphate Response Element Binding protein Binding Protein, also known as CREBBP or CBP or KAT3A, is a coactivator encoded by the CREBBP gene in humans, located on chromosome 16p13.3. CBP had intrinsic acetyltransferase functions; it is able to add acetyl groups to both transcription factors as well as histone lysines, the latter of which has been shown to alter chromatin structure making genes more accessible for transcription. This relatively unique acetyltransferase activity is also seen in another transcription enzyme, EP300 (p300). Together, they are known as the p300-CBP coactivator family and are known to associate with more than 16,000 genes in humans; however, while these proteins share many structural features, emerging evidence suggests that these two co-activators may promote transcription of genes with different biological functions.

Histone acetylation and deacetylation are the processes by which the lysine residues within the N-terminal tail protruding from the histone core of the nucleosome are acetylated and deacetylated as part of gene regulation.
Chromatin remodeling is the dynamic modification of chromatin architecture to allow access of condensed genomic DNA to the regulatory transcription machinery proteins, and thereby control gene expression. Such remodeling is principally carried out by 1) covalent histone modifications by specific enzymes, e.g., histone acetyltransferases (HATs), deacetylases, methyltransferases, and kinases, and 2) ATP-dependent chromatin remodeling complexes which either move, eject or restructure nucleosomes. Besides actively regulating gene expression, dynamic remodeling of chromatin imparts an epigenetic regulatory role in several key biological processes, egg cells DNA replication and repair; apoptosis; chromosome segregation as well as development and pluripotency. Aberrations in chromatin remodeling proteins are found to be associated with human diseases, including cancer. Targeting chromatin remodeling pathways is currently evolving as a major therapeutic strategy in the treatment of several cancers.

Probable global transcription activator SNF2L2 is a protein that in humans is encoded by the SMARCA2 gene.

SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 is a protein that in humans is encoded by the SMARCA5 gene.

Chromodomain-helicase-DNA-binding protein 3 is an enzyme that in humans is encoded by the CHD3 gene.
Tyrosine-protein kinase, or Bromodomain adjacent to zinc finger domain, 1B (BAZ1B) is an enzyme that in humans is encoded by the BAZ1B gene.

Probable global transcription activator SNF2L1 is a protein that in humans is encoded by the SMARCA1 gene.
Bromodomain adjacent to zinc finger domain protein 1A is a protein that in humans is encoded by the BAZ1A gene.
ISWI or imitation SWI is a protein found in the common fruit fly. It is the first ATPase subunit which has been isolated in the ISWI chromatin remodeling family. This protein presents high level of similarity to the SWI/SNF chromatin remodeling family in the ATPase domain. Outside the ATPase domain ISWI loses the similarity with the member of the SWI/SNF family, possessing a SANT domain instead of the bromodomain. The protein ISWI can interact with several proteins giving three different chromatin-remodeling complexes in Drosophila melanogaster: NURF, CHRAC and ACF.

In biochemistry, the KIX domain (kinase-inducible domain (KID) interacting domain) or CREB binding domain is a protein domain of the eukaryotic transcriptional coactivators CBP and P300. It serves as a docking site for the formation of heterodimers between the coactivator and specific transcription factors. Structurally, the KIX domain is a globular domain consisting of three α-helices and two short 310-helices.
In the field of molecular biology, the Mi-2/NuRDcomplex, is a group of associated proteins with both ATP-dependent chromatin remodeling and histone deacetylase activities. As of 2007, Mi-2/NuRD was the only known protein complex that couples chromatin remodeling ATPase and chromatin deacetylation enzymatic functions.

Memory is commonly referred to as the ability to encode, store, retain and subsequently recall information and past experiences in the human brain. This process involves many proteins, one of which is the Histone-binding protein RbAp48, encoded by the RBBP4 gene in humans.
The INO80 subfamily of chromatin remodeling complexes are ATPases, and includes the INO80 and SWR1 complexes.
Chromodomain helicase DNA-binding (CHD) proteins is a subfamily of ATP-dependent chromatin remodeling complexes (remodelers). All remodelers fall under the umbrella of RNA/DNA helicase superfamily 2. In yeast, CHD complexes are primarily responsible for nucleosome assembly and organization. These complexes play an additional role in multicellular eukaryotes, assisting in chromatin access and nucleosome editing.
Robert E. Kingston is an American biochemist who studies the functional and regulatory role nucleosomes play in gene expression, specifically during early development. After receiving his PhD (1981) and completing post-doctoral research, Kingston became an assistant professor at Massachusetts General Hospital (1985), where he started a research laboratory focused on understanding chromatin's structure with regards to transcriptional regulation. As a Harvard graduate himself, Kingston has served his alma mater through his leadership. He was the head of the Harvard's Biological and Biomedical Sciences PhD program from 2004 to 2007, the chair of the Molecular Biology Department at Massachusetts General Hospital from 2005 to present day, the vice-chair of the Department of Genetics at Harvard Medical School, and the chair of the executive committee on research at Massachusetts General Hospital from 2012 to 2015. In addition to being a professor of genetics at Harvard Medical School, Kingston frequently organizes conferences and performs editorials on his research interests. In 2016, he was elected by his peers to be a member of the National Academy of Sciences.
References
- ↑ Bozhenok L, Wade PA, Varga-Weisz P (2002). "WSTF-ISWI chromatin remodeling complex targets heterochromatic replication foci". EMBO J. 21 (9): 2231–41. doi:10.1093/emboj/21.9.2231. PMC 125993 . PMID 11980720.
- ↑ Ito T, Levenstein ME, Fyodorov DV, Kutach AK, Kobayashi R, Kadonaga JT (June 1999). "ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly". Genes Dev. 13 (12): 1529–39. doi:10.1101/gad.13.12.1529. PMC 316812 . PMID 10385622.
- ↑ Fyodorov DV, Kadonaga JT (September 2002). "Binding of Acf1 to DNA involves a WAC motif and is important for ACF-mediated chromatin assembly". Mol. Cell. Biol. 22 (18): 6344–53. doi:10.1128/mcb.22.18.6344-6353.2002. PMC 135643 . PMID 12192034.