| WWE protein domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Solution structure of WWE domain in BAB28015 | |||||||||
| Identifiers | |||||||||
| Symbol | WWE | ||||||||
| Pfam | PF02825 | ||||||||
| InterPro | IPR004170 | ||||||||
| |||||||||
| WWE protein domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Solution structure of WWE domain in BAB28015 | |||||||||
| Identifiers | |||||||||
| Symbol | WWE | ||||||||
| Pfam | PF02825 | ||||||||
| InterPro | IPR004170 | ||||||||
| |||||||||
The WWE domains occur in two functional classes of proteins, namely those associated with ubiquitination and those associated with poly-ADP ribosylation (PARP). Hence, WWE domains hold an important function in signal transduction, protein degradation, DNA repair and apoptosis. [1]
The WWE domain is named after three of its conserved residues, W and E residues (tryptophans and glutamate respectively), and is predicted to mediate specific protein-protein interactions in ubiquitin and ADP ribose conjugation systems (Poly ADP ribose polymerase). This domain is found as a tandem repeat at the N-terminal of Deltex, a cytosolic effector of Notch signalling thought to bind the N-terminal of the Notch receptor. [2] It is also found as an interaction module in protein ubiquination and ADP ribosylation proteins. [1]
Within each WWE module, the residues form two similar structures vital to its stability. The two WWE modules interact and form a large cleft suitable for binding of extended polypeptides. The two WWE modules adopt compact structures mostly composed of beta strands, with a single three turn alpha helix in both modules and an additional short helical segment in the second WWE module. The two WWE modules hold a two-fold rotation axis. [2]

Ubiquitin is a small regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ubiquitously. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Four genes in the human genome code for ubiquitin: UBB, UBC, UBA52 and RPS27A.
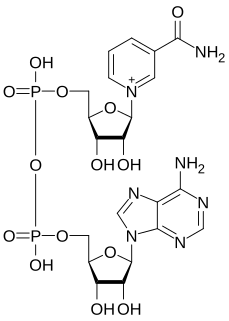
Nicotinamide adenine dinucleotide (NAD) is a coenzyme central to metabolism. Found in all living cells, NAD is called a dinucleotide because it consists of two nucleotides joined through their phosphate groups. One nucleotide contains an adenine nucleobase and the other nicotinamide. NAD exists in two forms: an oxidized and reduced form, abbreviated as NAD+ and NADH (H for hydrogen), respectively.

Poly (ADP-ribose) polymerase (PARP) is a family of proteins involved in a number of cellular processes such as DNA repair, genomic stability, and programmed cell death.

ADP ribosylation factors (ARFs) are members of the ARF family of GTP-binding proteins of the Ras superfamily. ARF family proteins are ubiquitous in eukaryotic cells, and six highly conserved members of the family have been identified in mammalian cells. Although ARFs are soluble, they generally associate with membranes because of N-terminus myristoylation. They function as regulators of vesicular traffic and actin remodelling.
The formyl peptide receptors (FPR) belong to a class of G protein-coupled receptors involved in chemotaxis. In humans, there are three formyl peptide receptor isoforms, each encoded by a separate gene that are named FPR1, FPR2, and FPR3. These receptors were originally identified by their ability to bind N-formyl peptides such as N-formylmethionine produced by the degradation of either bacterial or host cells. Hence formyl peptide receptors are involved in mediating immune cell response to infection. These receptors may also act to suppress the immune system under certain conditions. The close phylogenetic relation of signaling in chemotaxis and olfaction was recently proved by detection formyl peptide receptor like proteins as a distinct family of vomeronasal organ chemosensors in mice.

Histone-modifying enzymes are enzymes involved in the modification of histone substrates after protein translation and affect cellular processes including gene expression. To safely store the eukaryotic genome, DNA is wrapped around four core histone proteins, which then join to form nucleosomes. These nucleosomes further fold together into highly condensed chromatin, which renders the organism's genetic material far less accessible to the factors required for gene transcription, DNA replication, recombination and repair. Subsequently, eukaryotic organisms have developed intricate mechanisms to overcome this repressive barrier imposed by the chromatin through histone modification, a type of post-translational modification which typically involves covalently attaching certain groups to histone residues. Once added to the histone, these groups elicit either a loose and open histone conformation, euchromatin, or a tight and closed histone conformation, heterochromatin. Euchromatin marks active transcription and gene expression, as the light packing of histones in this way allows entry for proteins involved in the transcription process. As such, the tightly packed heterochromatin marks the absence of current gene expression.

ADP-ribosylation is the addition of one or more ADP-ribose moieties to a protein. It is a reversible post-translational modification that is involved in many cellular processes, including cell signaling, DNA repair, gene regulation and apoptosis. Improper ADP-ribosylation has been implicated in some forms of cancer. It is also the basis for the toxicity of bacterial compounds such as cholera toxin, diphtheria toxin, and others.

Protein C-ets-1 is a protein that in humans is encoded by the ETS1 gene. The protein encoded by this gene belongs to the ETS family of transcription factors.

Poly [ADP-ribose] polymerase 1 (PARP-1) also known as NAD+ ADP-ribosyltransferase 1 or poly[ADP-ribose] synthase 1 is an enzyme that in humans is encoded by the PARP1 gene. It is the most abundant of the PARP family of enzymes, accounting for 90% of the NAD+ used by the family. PARP1 is mostly present in cell nucleus, but cytosolic fraction of this protein was also reported.

ADP-ribosylation factor 1 is a protein that in humans is encoded by the ARF1 gene.

ADP-ribosylation factor-binding protein GGA1 is a protein that in humans is encoded by the GGA1 gene.

ADP-ribosylation factor-binding protein GGA3 is a protein that in humans is encoded by the GGA3 gene.

ADP-ribose diphosphatase (EC 3.6.1.13) is an enzyme that catalyzes a hydrolysis reaction in which water nucleophilically attacks ADP-ribose to produce AMP and D-ribose 5-phosphate. Enzyme hydrolysis occurs by the breakage of a phosphoanhydride bond and is dependent on Mg2+ ions that are held in complex by the enzyme.

The EGF-like domain is an evolutionary conserved protein domain, which derives its name from the epidermal growth factor where it was first described. It comprises about 30 to 40 amino-acid residues and has been found in a large number of mostly animal proteins. Most occurrences of the EGF-like domain are found in the extracellular domain of membrane-bound proteins or in proteins known to be secreted. An exception to this is the prostaglandin-endoperoxide synthase. The EGF-like domain includes 6 cysteine residues which in the epidermal growth factor have been shown to form 3 disulfide bonds. The structures of 4-disulfide EGF-domains have been solved from the laminin and integrin proteins. The main structure of EGF-like domains is a two-stranded β-sheet followed by a loop to a short C-terminal, two-stranded β-sheet. These two β-sheets are usually denoted as the major (N-terminal) and minor (C-terminal) sheets. EGF-like domains frequently occur in numerous tandem copies in proteins: these repeats typically fold together to form a single, linear solenoid domain block as a functional unit.

Histidine kinases (HK) are multifunctional, and in non-animal kingdoms, typically transmembrane, proteins of the transferase class of enzymes that play a role in signal transduction across the cellular membrane. The vast majority of HKs are homodimers that exhibit autokinase, phosphotransfer, and phosphatase activity. HKs can act as cellular receptors for signaling molecules in a way analogous to tyrosine kinase receptors (RTK). Multifunctional receptor molecules such as HKs and RTKs typically have portions on the outside of the cell that bind to hormone- or growth factor-like molecules, portions that span the cell membrane, and portions within the cell that contain the enzymatic activity. In addition to kinase activity, the intracellular domains typically have regions that bind to a secondary effector molecule or complex of molecules that further propagate signal transduction within the cell. Distinct from other classes of protein kinases, HKs are usually parts of a two-component signal transduction mechanisms in which HK transfers a phosphate group from ATP to a histidine residue within the kinase, and then to an aspartate residue on the receiver domain of a response regulator protein. More recently, the widespread existence of protein histidine phosphorylation distinct from that of two-component histidine kinases has been recognised in human cells. In marked contrast to Ser, Thr and Tyr phosphorylation, the analysis of phosphorylated Histidine using standard biochemical and mass spectrometric approaches is much more challenging, and special procedures and separation techniques are required for their preservation alongside classical Ser, Thr and Tyr phosphorylation on proteins isolated from human cells.
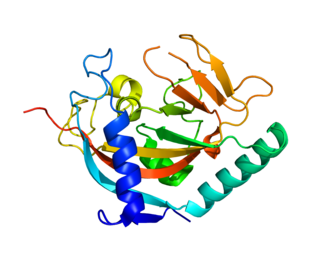
Tankyrase, also known as tankyrase 1, is an enzyme that in humans is encoded by the TNKS gene. It inhibits the binding of TERF1 to telomeric DNA. Tankyrase attracts substantial interest in cancer research through its interaction with AXIN1 and AXIN2, which are negative regulators of pro-oncogenic β-catenin signaling. Importantly, activity in the β-catenin destruction complex can be increased by tankyrase inhibitors and thus such inhibitors are a potential therapeutic option to reduce the growth of β-catenin-dependent cancers.

Poly [ADP-ribose] polymerase 2 is an enzyme that in humans is encoded by the PARP2 gene. It is one of the PARP family of enzymes.
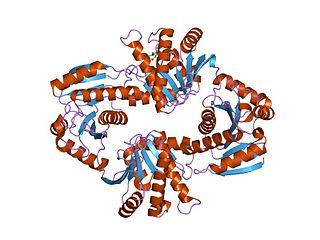
In molecular biology, the Macro domain or A1pp domain is a module of about 180 amino acids which can bind ADP-ribose, an NAD metabolite, or related ligands. Binding to ADP-ribose can be either covalent or non-covalent: in certain cases it is believed to bind non-covalently, while in other cases it appears to bind both non-covalently through a zinc finger motif, and covalently through a separate region of the protein.
Parthanatos is a form of programmed cell death that is distinct from other cell death processes such as necrosis and apoptosis. While necrosis is caused by acute cell injury resulting in traumatic cell death and apoptosis is a highly controlled process signalled by apoptotic intracellular signals, parthanatos is caused by the accumulation of Poly(ADP ribose) (PAR) and the nuclear translocation of apoptosis-inducing factor (AIF) from mitochondria. Parthanatos is also known as PARP-1 dependent cell death. PARP-1 mediates parthanatos when it is over-activated in response to extreme genomic stress and synthesizes PAR which causes nuclear translocation of AIF. Parthanatos is involved in diseases that afflict hundreds of millions of people worldwide. Well known diseases involving parthanatos include Parkinson's disease, stroke, heart attack, and diabetes. It also has potential use as a treatment for ameliorating disease and various medical conditions such as diabetes and obesity.
Ted M. Dawson is an American neurologist and neuroscientist. He is the Leonard and Madlyn Abramson Professor in Neurodegenerative Diseases and Director of the Institute for Cell Engineering at Johns Hopkins University School of Medicine. He has joint appointments in the Department of Neurology, Neuroscience and Department of Pharmacology and Molecular Sciences.