
β-galactosidase, also called lactase, beta-gal or β-gal, is a family of glycoside hydrolase enzymes that catalyzes the hydrolysis of β-galactosides into monosaccharides through the breaking of a glycosidic bond. β-galactosides include carbohydrates containing galactose where the glycosidic bond lies above the galactose molecule. Substrates of different β-galactosidases include ganglioside GM1, lactosylceramides, lactose, and various glycoproteins.

The nucleoid is an irregularly shaped region within the prokaryotic cell that contains all or most of the genetic material. The chromosome of a prokaryote is circular, and its length is very large compared to the cell dimensions, so it needs to be compacted in order to fit. In contrast to the nucleus of a eukaryotic cell, it is not surrounded by a nuclear membrane. Instead, the nucleoid forms by condensation and functional arrangement with the help of chromosomal architectural proteins and RNA molecules as well as DNA supercoiling. The length of a genome widely varies and a cell may contain multiple copies of it.

Transfer-messenger RNA is a bacterial RNA molecule with dual tRNA-like and messenger RNA-like properties. The tmRNA forms a ribonucleoprotein complex (tmRNP) together with Small Protein B (SmpB), Elongation Factor Tu (EF-Tu), and ribosomal protein S1. In trans-translation, tmRNA and its associated proteins bind to bacterial ribosomes which have stalled in the middle of protein biosynthesis, for example when reaching the end of a messenger RNA which has lost its stop codon. The tmRNA is remarkably versatile: it recycles the stalled ribosome, adds a proteolysis-inducing tag to the unfinished polypeptide, and facilitates the degradation of the aberrant messenger RNA. In the majority of bacteria these functions are carried out by standard one-piece tmRNAs. In other bacterial species, a permuted ssrA gene produces a two-piece tmRNA in which two separate RNA chains are joined by base-pairing.

EF-Tu is a prokaryotic elongation factor responsible for catalyzing the binding of an aminoacyl-tRNA (aa-tRNA) to the ribosome. It is a G-protein, and facilitates the selection and binding of an aa-tRNA to the A-site of the ribosome. As a reflection of its crucial role in translation, EF-Tu is one of the most abundant and highly conserved proteins in prokaryotes. It is found in eukaryotic mitochrondria as TUFM.

The lipocalins are a family of proteins which transport small hydrophobic molecules such as steroids, bilins, retinoids, and lipids and most lipocalins are also able to bind to complexed iron as well as heme. They share limited regions of sequence homology and a common tertiary structure architecture. This is an eight stranded antiparallel beta barrel with a repeated + 1 topology enclosing an internal ligand binding site.

Maltose-binding protein (MBP) is a part of the maltose/maltodextrin system of Escherichia coli, which is responsible for the uptake and efficient catabolism of maltodextrins. It is a complex regulatory and transport system involving many proteins and protein complexes. MBP has an approximate molecular mass of 42.5 kilodaltons.

4-hydroxyphenylacetate 3-monooxygenase (EC 1.14.14.9) is an enzyme that catalyzes the chemical reaction

In enzymology, a phenylalanine-tRNA ligase is an enzyme that catalyzes the chemical reaction
Tyrosine—tRNA ligase, also known as tyrosyl-tRNA synthetase is an enzyme that is encoded by the gene YARS. Tyrosine—tRNA ligase catalyzes the chemical reaction

General bacterial porins are a family of porin proteins from the outer membranes of Gram-negative bacteria. The porins act as molecular filters for hydrophilic compounds. They are responsible for the 'molecular sieve' properties of the outer membrane. Porins form large water-filled channels which allow the diffusion of hydrophilic molecules into the periplasmic space. Some porins form general diffusion channels that allow any solute up to a certain size to cross the membrane, while other porins are specific for one particular solute and contain a binding site for that solute inside the pores. As porins are the major outer membrane proteins, they also serve as receptor sites for the binding of phages and bacteriocins.

Outer membrane receptors, also known as TonB-dependent receptors, are a family of beta barrel proteins named for their localization in the outer membrane of gram-negative bacteria. TonB complexes sense signals from the outside of bacterial cells and transmit them into the cytoplasm, leading to transcriptional activation of target genes. TonB-dependent receptors in gram-negative bacteria are associated with the uptake and transport of large substrates such as iron siderophore complexes and vitamin B12.

The fimbrial usher protein is involved in biogenesis of the pilus in Gram-negative bacteria. The biogenesis of some fimbriae requires a two-component assembly and transport system which is composed of a periplasmic chaperone and a pore-forming outer membrane protein which has been termed a molecular 'usher'; this is the chaperone-usher pathway.
Aminoacyl-tRNA synthetases, class II is a family of proteins. These proteins catalyse the attachment of an amino acid to its cognate transfer RNA molecule in a highly specific two-step reaction. These proteins differ widely in size and oligomeric state, and have a limited sequence homology.
The aminoacyl-tRNA synthetases catalyse the attachment of an amino acid to its cognate transfer RNA molecule in a highly specific two-step reaction. These proteins differ widely in size and oligomeric state, and have limited sequence homology. The 20 aminoacyl-tRNA synthetases are divided into two classes, I and II. Class I aminoacyl-tRNA synthetases contain a characteristic Rossmann fold catalytic domain and are mostly monomeric. Class II aminoacyl-tRNA synthetases share an anti-parallel beta-sheet fold flanked by alpha-helices, and are mostly dimeric or multimeric, containing at least three conserved regions. However, tRNA binding involves an alpha-helical structure that is conserved between class I and class II synthetases. In reactions catalysed by the class I aminoacyl-tRNA synthetases, the aminoacyl group is coupled to the 2'-hydroxyl of the tRNA, while, in class II reactions, the 3'-hydroxyl site is preferred. The synthetases specific for arginine, cysteine, glutamic acid, glutamine, isoleucine, leucine, methionine, tyrosine, tryptophan and valine belong to class I synthetases; these synthetases are further divided into three subclasses, a, b and c, according to sequence homology. The synthetases specific for alanine, asparagine, aspartic acid, glycine, histidine, lysine, phenylalanine, proline, serine, and threonine belong to class-II synthetases.
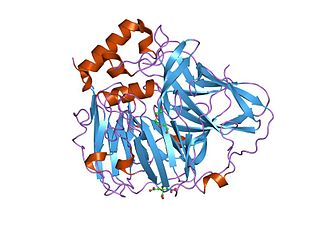
In molecular biology, multicopper oxidases are enzymes which oxidise their substrate by accepting electrons at a mononuclear copper centre and transferring them to a trinuclear copper centre; dioxygen binds to the trinuclear centre and, following the transfer of four electrons, is reduced to two molecules of water. There are three spectroscopically different copper centres found in multicopper oxidases: type 1, type 2 and type 3. Multicopper oxidases consist of 2, 3 or 6 of these homologous domains, which also share homology with the cupredoxins azurin and plastocyanin. Structurally, these domains consist of a cupredoxin-like fold, a beta-sandwich consisting of 7 strands in 2 beta-sheets, arranged in a Greek-key beta-barrel. Multicopper oxidases include:

DsbC is a prokaryotic disulfide bond isomerase. The formation of native disulfide bonds play an important role in the proper folding of proteins and stabilize tertiary structures of the protein. DsbC is one of 6 proteins in the Dsb family in prokaryotes. The other proteins are DsbA, DsbB, DsbD, DsbE and DsbG. These enzymes work in tandem with each other to form disulfide bonds during the expression of proteins. DsbC and DsbG act as proofreaders of the disulfide bonds that are formed. They break non-native disulfide bonds that were formed and act as chaperones for the formation of native disulfide bonds. The isomerization of disulfide bonds occurs in the periplasm.

EF-P is an essential protein that in bacteria stimulates the formation of the first peptide bonds in protein synthesis. Studies show that EF-P prevents ribosomes from stalling during the synthesis of proteins containing consecutive prolines. EF-P binds to a site located between the binding site for the peptidyl tRNA and the exiting tRNA. It spans both ribosomal subunits with its amino-terminal domain positioned adjacent to the aminoacyl acceptor stem and its carboxyl-terminal domain positioned next to the anticodon stem-loop of the P site-bound initiator tRNA. The EF-P protein shape and size is very similar to a tRNA and interacts with the ribosome via the exit “E” site on the 30S subunit and the peptidyl-transferase center (PTC) of the 50S subunit. EF-P is a translation aspect of an unknown function, therefore It probably functions indirectly by altering the affinity of the ribosome for aminoacyl-tRNA, thus increasing their reactivity as acceptors for peptidyl transferase.

In molecular biology, the isocitrate/isopropylmalate dehydrogenase family is a protein family consisting of the evolutionary related enzymes isocitrate dehydrogenase, 3-isopropylmalate dehydrogenase and tartrate dehydrogenase.

In molecular biology, ZinT is a family of protein domains found in prokaryotes. The domain contains a single binding site that can accommodate a divalent cation, with a geometry suggestive of zinc binding. This family was first thought to be part of the bacterial response to a toxic heavy metal cadmium by binding to the metal to ensure its elimination; however, more recent work has suggested a role in zinc homeostasis.

ZnuABC is a high-affinity transporter specialized for transporting zinc ions as part of a system for metal ion homeostasis in bacteria. The complex is a member of the ATP-binding cassette (ABC) transporter protein family. The transporter contains three protein components:

















