
Ion channels are pore-forming membrane proteins that allow ions to pass through the channel pore. Their functions include establishing a resting membrane potential, shaping action potentials and other electrical signals by gating the flow of ions across the cell membrane, controlling the flow of ions across secretory and epithelial cells, and regulating cell volume. Ion channels are present in the membranes of all cells. Ion channels are one of the two classes of ionophoric proteins, the other being ion transporters.

An olfactory receptor neuron (ORN), also called an olfactory sensory neuron (OSN), is a sensory neuron within the olfactory system.
Calcium release-activated channels (CRAC) are specialized plasma membrane Ca2+ ion channels. When calcium ions (Ca2+) are depleted from the endoplasmic reticulum (a major store of Ca2+) of mammalian cells, the CRAC channel is activated to slowly replenish the level of calcium in the endoplasmic reticulum. The Ca2+ Release-activated Ca2+ (CRAC) Channel (CRAC-C) Family (TC# 1.A.52) is a member of the Cation Diffusion Facilitator (CDF) Superfamily. These proteins typically have between 4 and 6 transmembrane α-helical spanners (TMSs). The 4 TMS CRAC channels arose by loss of 2TMSs from 6TMS CDF carriers, an example of 'reverse' evolution'.
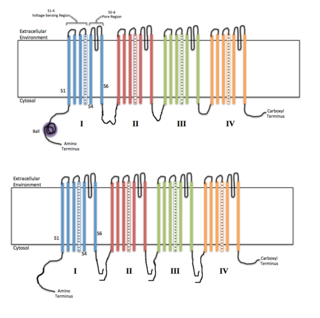
Voltage-gated ion channels are a class of transmembrane proteins that form ion channels that are activated by changes in the electrical membrane potential near the channel. The membrane potential alters the conformation of the channel proteins, regulating their opening and closing. Cell membranes are generally impermeable to ions, thus they must diffuse through the membrane through transmembrane protein channels. They have a crucial role in excitable cells such as neuronal and muscle tissues, allowing a rapid and co-ordinated depolarization in response to triggering voltage change. Found along the axon and at the synapse, voltage-gated ion channels directionally propagate electrical signals. Voltage-gated ion-channels are usually ion-specific, and channels specific to sodium (Na+), potassium (K+), calcium (Ca2+), and chloride (Cl−) ions have been identified. The opening and closing of the channels are triggered by changing ion concentration, and hence charge gradient, between the sides of the cell membrane.

Chloride channels are a superfamily of poorly understood ion channels specific for chloride. These channels may conduct many different ions, but are named for chloride because its concentration in vivo is much higher than other anions. Several families of voltage-gated channels and ligand-gated channels have been characterized in humans.
Calcium-activated potassium channels are potassium channels gated by calcium, or that are structurally or phylogenetically related to calcium gated channels. They were first discovered in 1958 by Gardos who saw that Calcium levels inside of a cell could affect the permeability of potassium through that cell membrane. Then in 1970, Meech was the first to observe that intracellular calcium could trigger potassium currents. In humans they are divided into three subtypes: large conductance or BK channels, which have very high conductance which range from 100 to 300 pS, intermediate conductance or IK channels, with intermediate conductance ranging from 25 to 100 pS, and small conductance or SK channels with small conductances from 2-25 pS.

Stromal interaction molecule 1 is a protein that in humans is encoded by the STIM1 gene. STIM1 has a single transmembrane domain, and is localized to the endoplasmic reticulum, and to a lesser extent to the plasma membrane.

The CLCN5 gene encodes the chloride channel Cl-/H+ exchanger ClC-5. ClC-5 is mainly expressed in the kidney, in particular in proximal tubules where it participates to the uptake of albumin and low-molecular-weight proteins, which is one of the principal physiological role of proximal tubular cells. Mutations in the CLCN5 gene cause an X-linked recessive nephropathy named Dent disease characterized by excessive urinary loss of low-molecular-weight proteins and of calcium (hypercalciuria), nephrocalcinosis and nephrolithiasis.

Bestrophin-1 (Best1) is a protein that, in humans, is encoded by the BEST1 gene.

Potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4, also known as KCNN4, is a human gene encoding the KCa3.1 protein.

Chloride channel accessory 1 is a protein that in humans is encoded by the CLCA1 gene.

Calcium channel, voltage-dependent, L type, alpha 1D subunit is a protein that in humans is encoded by the CACNA1D gene. Cav1.3 channels belong to the Cav1 family, which form L-type calcium currents and are sensitive to selective inhibition by dihydropyridines (DHP).

Chloride channel accessory 2 is a protein that in humans is encoded by the CLCA2 gene.

Chloride channel accessory 3, also known as CLCA3, is a protein which in humans is encoded by the CLCA3P pseudogene. The protein encoded by this gene is a chloride channel. According to the HGNC, this protein is not expressed in humans but is in certain other species such as mouse. However, some conflicting reports state that human cells produce and glycosylate this protein.

Anoctamin-1 (ANO1) also known as Transmembrane member 16A (TMEM16A) is a protein that, in humans, is encoded by the ANO1 gene. Anoctamin-1 is a voltage-gated calcium-activated anion channel, which acts as a chloride channel and a bicarbonate channel. additionally Anoctamin-1 is apical iodide channel. It is expressed in smooth muscle, epithelial cells, vomeronasal neurons, olfactory sustentacular cells, and is highly expressed in human interstitial cells of Cajal (ICC) throughout the gastrointestinal tract.
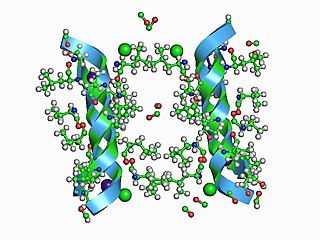
In electrophysiology, the term gating refers to the opening (activation) or closing of ion channels. This change in conformation is a response to changes in transmembrane voltage.
ANO3 is a gene that in humans is located on chromosome 11 and encodes the protein anoctamin 3. It belongs to a family of genes (ANO1–ANO10) that appear to encode calcium-activated chloride channels.

Anoctamin 6 is a protein that in humans is encoded by the ANO6 gene.

Anoctamin 5 (ANO5) is a protein that in humans is encoded by the ANO5 gene.












