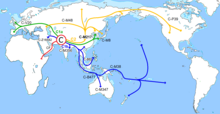
In human genetics, Haplogroup J-M172 or J2 is a Y-chromosome haplogroup which is a subclade (branch) of haplogroup J-M304. Haplogroup J-M172 is common in modern populations in Western Asia, Central Asia, South Asia, Southern Europe, Northwestern Iran and North Africa. It is thought that J-M172 may have originated between the Caucasus, Anatolia and/or Western Iran.
Haplogroup D1 or D-M174 is a subclade of haplogroup D-CTS3946. This male haplogroup is found primarily in East Asia, Magar-ethnic Nepal and the Andaman Islands. It is also found regularly with lower frequency in Central Asia and Mainland Southeast Asia, and, more rarely, in Europe and the Middle East.
Haplogroup E-M96 is a human Y-chromosome DNA haplogroup. It is one of the two main branches of the older and ancestral haplogroup DE, the other main branch being haplogroup D. The E-M96 clade is divided into two main subclades: the more common E-P147, and the less common E-M75.
Haplogroup E-V38, also known as E1b1a-V38, is a human Y-chromosome DNA haplogroup. E-V38 is primarily distributed in sub-Saharan Africa. E-V38 has two basal branches, E-M329 and E-M2. E-M329 is a subclade mostly found in East Africa. E-M2 is the predominant subclade in West Africa, Central Africa, Southern Africa, and the region of African Great Lakes; it also occurs at moderate frequencies in some parts of North Africa, West Asia, and Southern Europe.

Haplogroup L-M20 is a human Y-DNA haplogroup, which is defined by SNPs M11, M20, M61 and M185. As a secondary descendant of haplogroup K and a primary branch of haplogroup LT, haplogroup L currently has the alternative phylogenetic name of K1a, and is a sibling of haplogroup T.

Haplogroup M, also known as M-P256 and Haplogroup K2b1b is a Y-chromosome DNA haplogroup. M-P256 is a descendant haplogroup of Haplogroup K2b1, and is believed to have first appeared between 32,000 and 47,000 years ago.
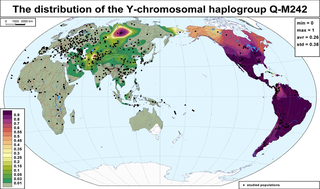
Haplogroup Q or Q-M242 is a Y-chromosome DNA haplogroup. It has one primary subclade, Haplogroup Q1 (L232/S432), which includes numerous subclades that have been sampled and identified in males among modern populations.

In human genetics, a human Y-chromosome DNA haplogroup is a haplogroup defined by mutations in the non-recombining portions of DNA from the male-specific Y chromosome. Many people within a haplogroup share similar numbers of short tandem repeats (STRs) and types of mutations called single-nucleotide polymorphisms (SNPs).
Haplogroup R, or R-M207, is a Y-chromosome DNA haplogroup. It is both numerous and widespread amongst modern populations.
In human genetics, Haplogroup O-M119 is a Y-chromosome DNA haplogroup. Haplogroup O-M119 is a descendant branch of haplogroup O-F265 also known as O1a, one of two extant primary subclades of Haplogroup O-M175. The same clade previously has been labeled as O-MSY2.2.

Haplogroup C-M217, also known as C2, is a Y-chromosome DNA haplogroup. It is the most frequently occurring branch of the wider Haplogroup C (M130). It is found mostly in Central Asia, Eastern Siberia and significant frequencies in parts of East Asia and Southeast Asia including some populations in the Caucasus, Middle East, South Asia, East Europe. It is found in a much more widespread areas with a low frequency of less than 2%.
Haplogroup DE is a human Y-chromosome DNA haplogroup. It is defined by the single nucleotide polymorphism (SNP) mutations, or UEPs, M1(YAP), M145(P205), M203, P144, P153, P165, P167, P183. DE is unique because it is distributed in several geographically distinct clusters. An immediate subclade, haplogroup D, is mainly found in East Asia, parts of Central Asia, and the Andaman Islands, but also sporadically in West Africa and West Asia. The other immediate subclade, haplogroup E, is common in Africa, and to a lesser extent the Middle East and southern Europe.

Haplogroup CT is a human Y chromosome haplogroup, defining one of the major paternal lineages of humanity.
Haplogroup CF, also known as CF-P143 and CT(xDE), is a human Y-chromosome DNA haplogroup. This paternal lineage is defined by the SNP P143. The clade's existence and distribution are inferred from the fact that haplogroups descended from CF include most human male lineages in Eurasia, Oceania and The Americas.

Haplogroup S-M230, also known as S1a1b, is a Y-chromosome DNA haplogroup. It is by far the most numerically significant subclade of Haplogroup S1a.
Haplogroup E-M75 is a human Y-chromosome DNA haplogroup. Along with haplogroup E-P147, it is one of the two main branches of the older haplogroup E-M96.
Haplogroup E-V68, also known as E1b1b1a, is a major human Y-chromosome DNA haplogroup found in North Africa, the Horn of Africa, Western Asia and Europe. It is a subclade of the larger and older haplogroup, known as E1b1b or E-M215. The E1b1b1a lineage is identified by the presence of a single nucleotide polymorphism (SNP) mutation on the Y chromosome, which is known as V68. It is a subject of discussion and study in genetics as well as genetic genealogy, archaeology, and historical linguistics.
Haplogroup E-P2, also known as E1b1, is a human Y-chromosome DNA haplogroup. E-P2 has two basal branches, E-V38 and E-M215. E-P2 had an ancient presence in East Africa and the Levant; presently, it is primarily distributed in Africa where it may have originated, and occurs at lower frequencies in the Middle East and Europe.
In human population genetics, Y-Chromosome haplogroups define the major lineages of direct paternal (male) lines back to a shared common ancestor in Africa. Men in the same haplogroup share a set of differences, or markers, on their Y-Chromosome, which distinguish them from men in other haplogroups. These UEPs, or markers used to define haplogroups, are SNP mutations. Y-Chromosome Haplogroups all form "family trees" or "phylogenies", with both branches or sub-clades diverging from a common haplogroup ancestor, and also with all haplogroups themselves linked into one family tree which traces back ultimately to the most recent shared male line ancestor of all men alive today, called in popular science Y Chromosome Adam.
Haplogroup C1 also known as C-F3393, is a major Y-chromosome haplogroup. It is one of two primary branches of the broader Haplogroup C, the other being C2.