Related Research Articles

An antibiotic is a type of antimicrobial substance active against bacteria. It is the most important type of antibacterial agent for fighting bacterial infections, and antibiotic medications are widely used in the treatment and prevention of such infections. They may either kill or inhibit the growth of bacteria. A limited number of antibiotics also possess antiprotozoal activity. Antibiotics are not effective against viruses such as the ones which cause the common cold or influenza; drugs which inhibit growth of viruses are termed antiviral drugs or antivirals rather than antibiotics. They are also not effective against fungi; drugs which inhibit growth of fungi are called antifungal drugs.

Antimicrobial resistance (AMR) occurs when microbes evolve mechanisms that protect them from the effects of antimicrobials. All classes of microbes can evolve resistance where the drugs are no longer effective. Fungi evolve antifungal resistance, viruses evolve antiviral resistance, protozoa evolve antiprotozoal resistance, and bacteria evolve antibiotic resistance. Together all of these come under the umbrella of antimicrobial resistance. Microbes resistant to multiple antimicrobials are called multidrug resistant (MDR) and are sometimes referred to as superbugs. Although antimicrobial resistance is a naturally occurring process, it is often the result of improper usage of the drugs and management of the infections.

Mycoplasma genitalium is a sexually transmitted, small and pathogenic bacterium that lives on the mucous epithelial cells of the urinary and genital tracts in humans. Medical reports published in 2007 and 2015 state that Mgen is becoming increasingly common. Resistance to multiple antibiotics, including the macrolide azithromycin, which until recently was the most reliable treatment, is becoming prevalent. The bacteria was first isolated from the urogenital tract of humans in 1981, and was eventually identified as a new species of Mycoplasma in 1983. It can cause negative health effects in men and women. It also increases the risk factor for HIV spread with higher occurrences in those previously treated with the azithromycin antibiotics.

Klebsiella pneumoniae is a Gram-negative, non-motile, encapsulated, lactose-fermenting, facultative anaerobic, rod-shaped bacterium. It appears as a mucoid lactose fermenter on MacConkey agar.
Azithromycin, sold under the brand names Zithromax and Azasite, is an antibiotic medication used for the treatment of a number of bacterial infections. This includes middle ear infections, strep throat, pneumonia, traveler's diarrhea, and certain other intestinal infections. Along with other medications, it may also be used for malaria. It can be taken by mouth or intravenously.

A hospital-acquired infection, also known as a nosocomial infection, is an infection that is acquired in a hospital or other healthcare facility. To emphasize both hospital and nonhospital settings, it is sometimes instead called a healthcare-associated infection. Such an infection can be acquired in hospital, nursing home, rehabilitation facility, outpatient clinic, diagnostic laboratory or other clinical settings. A number of dynamic processes can bring contamination into operating rooms and other areas within nosocomial settings. Infection is spread to the susceptible patient in the clinical setting by various means. Healthcare staff also spread infection, in addition to contaminated equipment, bed linens, or air droplets. The infection can originate from the outside environment, another infected patient, staff that may be infected, or in some cases, the source of the infection cannot be determined. In some cases the microorganism originates from the patient's own skin microbiota, becoming opportunistic after surgery or other procedures that compromise the protective skin barrier. Though the patient may have contracted the infection from their own skin, the infection is still considered nosocomial since it develops in the health care setting. Nosocomial infection tends to lack evidence that it was present when the patient entered the healthcare setting, thus meaning it was acquired post-admission.

Colistin, also known as polymyxin E, is an antibiotic medication used as a last-resort treatment for multidrug-resistant Gram-negative infections including pneumonia. These may involve bacteria such as Pseudomonas aeruginosa, Klebsiella pneumoniae, or Acinetobacter. It comes in two forms: colistimethate sodium can be injected into a vein, injected into a muscle, or inhaled, and colistin sulfate is mainly applied to the skin or taken by mouth. Colistimethate sodium is a prodrug; it is produced by the reaction of colistin with formaldehyde and sodium bisulfite, which leads to the addition of a sulfomethyl group to the primary amines of colistin. Colistimethate sodium is less toxic than colistin when administered parenterally. In aqueous solutions it undergoes hydrolysis to form a complex mixture of partially sulfomethylated derivatives, as well as colistin. Resistance to colistin began to appear as of 2015.
Multiple drug resistance (MDR), multidrug resistance or multiresistance is antimicrobial resistance shown by a species of microorganism to at least one antimicrobial drug in three or more antimicrobial categories. Antimicrobial categories are classifications of antimicrobial agents based on their mode of action and specific to target organisms. The MDR types most threatening to public health are MDR bacteria that resist multiple antibiotics; other types include MDR viruses, parasites.

Carbapenems are a class of very effective antibiotic agents most commonly used for treatment of severe bacterial infections. This class of antibiotics is usually reserved for known or suspected multidrug-resistant (MDR) bacterial infections. Similar to penicillins and cephalosporins, carbapenems are members of the beta-lactam antibiotics drug class, which kill bacteria by binding to penicillin-binding proteins, thus inhibiting bacterial cell wall synthesis. However, these agents individually exhibit a broader spectrum of activity compared to most cephalosporins and penicillins. Furthermore, carbapenems are typically unaffected by emerging antibiotic resistance, even to other beta-lactams.

Acinetobacter baumannii is a typically short, almost round, rod-shaped (coccobacillus) Gram-negative bacterium. It is named after the bacteriologist Paul Baumann. It can be an opportunistic pathogen in humans, affecting people with compromised immune systems, and is becoming increasingly important as a hospital-derived (nosocomial) infection. While other species of the genus Acinetobacter are often found in soil samples, it is almost exclusively isolated from hospital environments. Although occasionally it has been found in environmental soil and water samples, its natural habitat is still not known.

NDM-1 is an enzyme that makes bacteria resistant to a broad range of beta-lactam antibiotics. These include the antibiotics of the carbapenem family, which are a mainstay for the treatment of antibiotic-resistant bacterial infections. The gene for NDM-1 is one member of a large gene family that encodes beta-lactamase enzymes called carbapenemases. Bacteria that produce carbapenemases are often referred to in the news media as "superbugs" because infections caused by them are difficult to treat. Such bacteria are usually sensitive only to polymyxins and tigecycline.
Multidrug resistant Gram-negative bacteria are a type of Gram-negative bacteria with resistance to multiple antibiotics. They can cause bacteria infections that pose a serious and rapidly emerging threat for hospitalized patients and especially patients in intensive care units. Infections caused by MDR strains are correlated with increased morbidity, mortality, and prolonged hospitalization. Thus, not only do these bacteria pose a threat to global public health, but also create a significant burden to healthcare systems.

Rifalazil is an antibiotic substance that kills bacterial cells by blocking off the β-subunit in RNA polymerase. Rifalazil is used as a treatment for many different diseases. The most common are Chlamydia infection, Clostridium difficile associated diarrhea (CDAD), and tuberculosis (TB). Using rifalazil and the effects that coincide with taking rifalazil for treating a bacterial disease vary from person to person, as does any drug put into the human body. Food interactions and genetic variation are a few causes for the variation in side effects from the use of rifalazil. Its development was terminated in 2013 due to severe side effects.
Carbapenem-resistant Enterobacteriaceae (CRE) or carbapenemase-producing Enterobacteriaceae (CPE) are Gram-negative bacteria that are resistant to the carbapenem class of antibiotics, considered the drugs of last resort for such infections. They are resistant because they produce an enzyme called a carbapenemase that disables the drug molecule. The resistance can vary from moderate to severe. Enterobacteriaceae are common commensals and infectious agents. Experts fear CRE as the new "superbug". The bacteria can kill up to half of patients who get bloodstream infections. Tom Frieden, former head of the Centers for Disease Control and Prevention has referred to CRE as "nightmare bacteria". Examples of enzymes found in certain types of CRE are KPC and NDM. KPC and NDM are enzymes that break down carbapenems and make them ineffective. Both of these enzymes, as well as the enzyme VIM have also been reported in Pseudomonas.

Clostridioides difficile is a bacterium that is well known for causing serious diarrheal infections, and may also cause colon cancer. It is known also as C. difficile, or C. diff, and is a Gram-positive species of spore-forming bacteria. Clostridioides spp. are anaerobic, motile bacteria, ubiquitous in nature and especially prevalent in soil. Its vegetative cells are rod-shaped, pleomorphic, and occur in pairs or short chains. Under the microscope, they appear as long, irregular cells with a bulge at their terminal ends. Under Gram staining, C. difficile cells are Gram-positive and show optimum growth on blood agar at human body temperatures in the absence of oxygen. C. difficile is catalase- and superoxide dismutase-negative, and produces up to three types of toxins: enterotoxin A, cytotoxin B and Clostridioides difficile transferase. Under stress conditions, the bacteria produce spores that are able to tolerate extreme conditions that the active bacteria cannot tolerate.

Ceftazidime/avibactam, sold under the brand name Avycaz among others, is a fixed-dose combination medication composed of ceftazidime, a cephalosporin antibiotic, and avibactam, a β-lactamase inhibitor. It is used to treat complicated intra-abdominal infections, urinary tract infections, and pneumonia. It is only recommended when other options are not appropriate. It is given by injection into a vein.
ESKAPE is an acronym comprising the scientific names of six highly virulent and antibiotic resistant bacterial pathogens including: Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp. The acronym is sometimes extended to ESKAPEE to include Escherichia coli. This group of Gram-positive and Gram-negative bacteria can evade or 'escape' commonly used antibiotics due to their increasing multi-drug resistance (MDR). As a result, throughout the world, they are the major cause of life-threatening nosocomial or hospital-acquired infections in immunocompromised and critically ill patients who are most at risk. P. aeruginosa and S. aureus are some of the most ubiquitous pathogens in biofilms found in healthcare. P. aeruginosa is a Gram-negative, rod-shaped bacterium, commonly found in the gut flora, soil, and water that can be spread directly or indirectly to patients in healthcare settings. The pathogen can also be spread in other locations through contamination, including surfaces, equipment, and hands. The opportunistic pathogen can cause hospitalized patients to have infections in the lungs, blood, urinary tract, and in other body regions after surgery. S. aureus is a Gram-positive, cocci-shaped bacterium, residing in the environment and on the skin and nose of many healthy individuals. The bacterium can cause skin and bone infections, pneumonia, and other types of potentially serious infections if it enters the body. S. aureus has also gained resistance to many antibiotic treatments, making healing difficult. Because of natural and unnatural selective pressures and factors, antibiotic resistance in bacteria usually emerges through genetic mutation or acquires antibiotic-resistant genes (ARGs) through horizontal gene transfer - a genetic exchange process by which antibiotic resistance can spread.
Cefiderocol, sold under the brand name Fetroja among others, is an antibiotic used to treat complicated urinary tract infections when no other options are available. It is indicated for the treatment of multi-drug-resistant Gram-negative bacteria including Pseudomonas aeruginosa. It is given by injection into a vein.
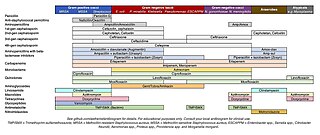
The antimicrobial spectrum of an antibiotic means the range of microorganisms it can kill or inhibit. Antibiotics can be divided into broad-spectrum antibiotics, extended-spectrum antibiotics and narrow-spectrum antibiotics based on their spectrum of activity. Detailedly, broad-spectrum antibiotics can kill or inhibit a wide range of microorganisms; extended-spectrum antibiotic can kill or inhibit Gram positive bacteria and some Gram negative bacteria; narrow-spectrum antibiotic can only kill or inhibit limited species of bacteria.

Multidrug-resistant bacteria are bacteria that are resistant to three or more classes of antimicrobial drugs. MDR bacteria have seen an increase in prevalence in recent years and pose serious risks to public health. MDR bacteria can be broken into 3 main categories: Gram-positive, Gram-negative, and other (acid-stain). These bacteria employ various adaptations to avoid or mitigate the damage done by antimicrobials. With increased access to modern medicine there has been a sharp increase in the amount of antibiotics consumed. Given the abundant use of antibiotics there has been a considerable increase in the evolution of antimicrobial resistance factors, now outpacing the development of new antibiotics.
References
- ↑ Baym M, Lieberman TD, Kelsic ED, Chait R, Gross R, Yelin I, Kishony R (2016-09-09). "Spatiotemporal microbial evolution on antibiotic landscapes". Science. 353 (6304): 1147–1151. Bibcode:2016Sci...353.1147B. doi:10.1126/science.aag0822. ISSN 0036-8075. PMC 5534434 . PMID 27609891.
- 1 2 Walsh TR, Weeks J, Livermore DM, Toleman MA (2011). "Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study". The Lancet Infectious Diseases. 11 (5): 355–362. doi:10.1016/s1473-3099(11)70059-7. PMID 21478057.
- ↑ Marilynn Marchione (13 September 2010). "New drug-resistant superbugs found in 3 states". Boston Globe.
- ↑ Madeleine White (21 August 2010). "Superbug detected in GTA". Toronto Star.
- ↑ Yuasa S (8 September 2010). "Japan confirms first case of superbug gene". The Boston Globe.
- ↑ Gerding DN, Johnson S, Peterson LR, Mulligan ME, Silva J (1995). "Clostridium difficile-associated diarrhea and colitis". Infect. Control Hosp. Epidemiol. 16 (8): 459–477. doi:10.1086/648363. PMID 7594392.
- ↑ McDonald LC (2005). "Clostridium difficile: responding to a new threat from an old enemy". Infect. Control Hosp. Epidemiol. 26 (8): 672–5. doi: 10.1086/502600 . PMID 16156321.
- 1 2 3 4 5 6 "Biggest Threats – Antibiotic/Antimicrobial Resistance – CDC". www.cdc.gov. Retrieved 2016-05-05.
- ↑ "CDC Press Releases". CDC. January 2016. Retrieved 2016-05-05.
- ↑ Baxter R, Ray GT, Fireman BH (January 2008). "Case-control study of antibiotic use and subsequent Clostridium difficile-associated diarrhea in hospitalized patients". Infection Control and Hospital Epidemiology. 29 (1): 44–50. doi:10.1086/524320. PMID 18171186. S2CID 39290661.
- ↑ Gifford AH, Kirkland KB (December 2006). "Risk factors for Clostridium difficile-associated diarrhea on an adult hematology-oncology ward". European Journal of Clinical Microbiology & Infectious Diseases. 25 (12): 751–5. doi:10.1007/s10096-006-0220-1. PMID 17072575. S2CID 23822514.
- ↑ Palmore TN, Sohn S, Malak SF, Eagan J, Sepkowitz KA (August 2005). "Risk factors for acquisition of Clostridium difficile-associated diarrhea among outpatients at a cancer hospital". Infection Control and Hospital Epidemiology. 26 (8): 680–4. doi:10.1086/502602. PMC 5612438 . PMID 16156323.
- ↑ Johnson S, Samore MH, Farrow KA, Killgore GE, Tenover FC, Lyras D, Rood JI, DeGirolami P, Baltch AL, Rafferty ME, Pear SM, Gerding DN (1999). "Epidemics of diarrhea caused by a clindamycin-resistant strain of Clostridium difficile in four hospitals". New England Journal of Medicine. 341 (23): 1645–1651. doi: 10.1056/NEJM199911253412203 . PMID 10572152.
- ↑ Loo VG, Poirier L, Miller MA, Oughton M, Libman MD, Michaud S, Bourgault AM, Nguyen T, Frenette C, Kelly M, Vibien A, Brassard P, Fenn S, Dewar K, Hudson TJ, Horn R, René P, Monczak Y, Dascal A (2005). "A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality". N Engl J Med. 353 (23): 2442–9. doi: 10.1056/NEJMoa051639 . PMID 16322602.
- ↑ Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, Fridkin SK (November 2008). National Healthcare Safety Network Team; Participating National Healthcare Safety Network Facilities. "NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007". Infect Control Hosp Epidemiol. 29 (11): 996–1011. doi:10.1086/591861. PMID 18947320. S2CID 205988392.
- ↑ Kristich CJ, Rice LB, Arias CA (2014-01-01). Gilmore MS, Clewell DB, Ike Y, Shankar N (eds.). Enterococcal Infection—Treatment and Antibiotic Resistance. Boston: Massachusetts Eye and Ear Infirmary. PMID 24649502.
- 1 2 "Antimicrobial Resistance Still Poses a Public Health Threat: A Conversation With Edward J. Septimus, MD, FIDSA, FACP, FSHEA, Clinical Professor of Internal Medicine at Texas A&M Health Science Center". Agency for Healthcare Research and Quality. 2013-04-17. Retrieved 2013-09-26.
- 1 2 LoBue P (2009). "Extensively drug-resistant tuberculosis". Current Opinion in Infectious Diseases. 22 (2): 167–73. doi:10.1097/QCO.0b013e3283229fab. PMID 19283912. S2CID 24995375.
- ↑ Brites D, Gagneux S (2013-04-17). "Co-evolution of Mycobacterium tuberculosis and Homo sapiens, Brites, D., & Gagneux, S. (2015)". Immunol Rev. 264 (1): 6–24. doi:10.1111/imr.12264. PMC 4339235 . PMID 25703549.
- ↑ Herzog H (1998). "History of Tuberculosis". Respiration. 65 (1): 5–15. doi:10.1159/000029220. PMID 9523361. S2CID 202645306.
- ↑ Gao Q, Li X (2010). "Transmission of MDR tuberculosis". Drug Discovery Today: Disease Mechanisms. 7: e61–e65. doi:10.1016/j.ddmec.2010.09.006.
- 1 2 Zainuddin ZF, Dale JW (1990). "Does Mycobacterium tuberculosis have plasmids?". Tubercle. 71 (1): 43–9. doi:10.1016/0041-3879(90)90060-l. PMID 2115217.
- ↑ Louw GE, Warren RM, Gey van Pittius NC, McEvoy CR, Van Helden PD, Victor TC (2009). "A Balancing Act: Efflux/Influx in Mycobacterial Drug Resistance". Antimicrobial Agents and Chemotherapy. 53 (8): 3181–9. doi:10.1128/AAC.01577-08. PMC 2715638 . PMID 19451293.
- ↑ Suneta S, Parkhouse A, Gillian D (24 April 2017). "Macrolide and quinolone-resistant Mycoplasma genitalium in a man with persistent urethritis: the tip of the British iceberg?". Sexually Transmitted Infections. 93 (8): 556–557. doi:10.1136/sextrans-2016-053077. PMID 28438948. S2CID 9178150 . Retrieved 6 October 2017.
- ↑ Yew HS, Anderson T, Coughlan E, Werno A (2011). "Induced macrolide resistance in Mycoplasma genitalium isolates from patients with recurrent nongonococcal urethritis". Journal of Clinical Microbiology. 49 (4): 1695–1696. doi:10.1128/JCM.02475-10. PMC 3122813 . PMID 21346049.
- ↑ Anagrius C, Loré B, Jensen JS, Coenye T (2013). "Treatment of Mycoplasma genitalium. Observations from a Swedish STD Clinic". PLOS ONE. 8 (4): e61481. Bibcode:2013PLoSO...861481A. doi: 10.1371/journal.pone.0061481 . PMC 3620223 . PMID 23593483.
- 1 2 3 Unemo M, Jensen JS (10 January 2017). "Antimicrobial-resistant sexually transmitted infections: gonorrhoea and Mycoplasma genitalium". Nature Reviews Urology. 14 (3): 139–125. doi:10.1038/nrurol.2016.268. PMID 28072403. S2CID 205521926.
- ↑ "Mycoplasma Genitalium Treatment Choices". www.theonlineclinic.co.uk.
- ↑ Jensen J, Cusini M, Gomberg M, Moi M (9 August 2016). "2016 European guideline on Mycoplasma genitalium infections". Journal of the European Academy of Dermatology and Venereology. 30 (10): 1650–1656. doi: 10.1111/jdv.13849 . PMID 27505296.
- 1 2 3 4 5 6 Murray CJ, Ikuta KS, Sharara F, Swetschinski L, Aguilar GR, Gray A, Han C, Bisignano C, Rao P, Wool E, Johnson SC (2022-01-19). "Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis". The Lancet. 399 (10325): 629–655. doi:10.1016/S0140-6736(21)02724-0. ISSN 0140-6736. PMC 8841637 . PMID 35065702.
- 1 2 Albrich WC, Monnet DL, Harbarth S (2004). "Antibiotic selection pressure and resistance in Streptococcus pneumoniae and Streptococcus pyogenes". Emerg. Infect. Dis. 10 (3): 514–7. doi:10.3201/eid1003.030252. PMC 3322805 . PMID 15109426.
- ↑ Hudson C, Bent Z, Meagher R, Williams K (June 7, 2014). "Resistance Determinants and Mobile Genetic Elements of an NDM-1-Encoding Klebsiella pneumoniae Strain". PLOS ONE. 9 (6): e99209. Bibcode:2014PLoSO...999209H. doi: 10.1371/journal.pone.0099209 . PMC 4048246 . PMID 24905728.
- ↑ Arnold RS, Thom KA, Sharma S, Phillips M, Kristie Johnson J, Morgan DJ (2011). "Emergence of Klebsiella pneumoniae Carbapenemase-Producing Bacteria". Southern Medical Journal. 104 (1): 40–5. doi:10.1097/SMJ.0b013e3181fd7d5a. PMC 3075864 . PMID 21119555.
- ↑ Davies J, Davies D (2010). "Origins and Evolution of Antibiotic Resistance". Microbiol Mol Biol Rev. 74 (3): 417–433. doi:10.1128/MMBR.00016-10. PMC 2937522 . PMID 20805405.
- 1 2 3 Adam M, Murali B, Glenn NO, Potter SS (2008). "Epigenetic inheritance based evolution of antibiotic resistance in bacteria". BMC Evol. Biol. 8 (1): 52. Bibcode:2008BMCEE...8...52A. doi: 10.1186/1471-2148-8-52 . PMC 2262874 . PMID 18282299.
- 1 2 Reardon S (21 December 2015). "Spread of antibiotic-resistance gene does not spell bacterial apocalypse — yet". Nature. doi:10.1038/nature.2015.19037. S2CID 182042290.
- ↑ Centers for Disease Control Prevention (CDC). (2004). "Acinetobacter baumannii infections among patients at military medical facilities treating injured U.S. service members, 2002–2004". MMWR Morb. Mortal. Wkly. Rep. 53 (45): 1063–6. PMID 15549020.
- ↑ "Medscape abstract on Acinetobacter baumannii: Acinetobacter baumannii: An Emerging Multidrug-resistant Threat".
membership only website
- ↑ Poole K (2004). "Efflux-mediated multiresistance in Gram-negative bacteria". Clinical Microbiology and Infection. 10 (1): 12–26. doi: 10.1111/j.1469-0691.2004.00763.x . PMID 14706082.
- ↑ Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK (2011). "Active Starvation Responses Mediate Antibiotic Tolerance in Biofilms and Nutrient-Limited Bacteria". Science. 334 (6058): 982–6. Bibcode:2011Sci...334..982N. doi:10.1126/science.1211037. PMC 4046891 . PMID 22096200.