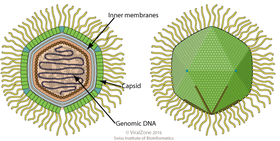
Mimivirus is a genus of giant viruses, in the family Mimiviridae. Amoeba serve as their natural hosts. This genus contains a single identified species named Acanthamoeba polyphaga mimivirus (APMV). It also refers to a group of phylogenetically related large viruses.
A satellite is a subviral agent that depends on the coinfection of a host cell with a helper virus for its replication. Satellites can be divided into two major classes: satellite viruses and satellite nucleic acids. Satellite viruses, which are most commonly associated with plants, are also found in mammals, arthropods, and bacteria. They encode structural proteins to enclose their genetic material, which are therefore distinct from the structural proteins of their helper viruses. Satellite nucleic acids, in contrast, do not encode their own structural proteins, but instead are encapsulated by proteins encoded by their helper viruses. The genomes of satellites range upward from 359 nucleotides in length for satellite tobacco ringspot virus RNA (STobRV).

Lake Pampulha is an artificial lake located in Pampulha, Belo Horizonte, Brazil. It is also the name of an administrative region of Belo Horizonte, and the name of one of 29 neighborhoods (bairros) within the administrative region of the same name. The lake was built in the early 1940s during the mayoralty of Juscelino Kubitschek, later president of Brazil from 1956 to 1961. Pampulha was created as a source of water for the city of Belo Horizonte, but quickly became polluted.

The mobilome is the entire set of mobile genetic elements in a genome. Mobilomes are found in eukaryotes, prokaryotes, and viruses. The compositions of mobilomes differ among lineages of life, with transposable elements being the major mobile elements in eukaryotes, and plasmids and prophages being the major types in prokaryotes. Virophages contribute to the viral mobilome.

Asfarviridae is a family of viruses, the best-studied of which is African swine fever virus. They are double-stranded DNA viruses.

Virophages are small, double-stranded DNA viral phages that require the co-infection of another virus. The co-infecting viruses are typically giant viruses. Virophages rely on the viral replication factory of the co-infecting giant virus for their own replication. One of the characteristics of virophages is that they have a parasitic relationship with the co-infecting virus. Their dependence upon the giant virus for replication often results in the deactivation of the giant viruses. The virophage may improve the recovery and survival of the host organism.

Rickettsia conorii is a Gram-negative, obligate intracellular bacterium of the genus Rickettsia that causes human disease called boutonneuse fever, Mediterranean spotted fever, Israeli tick typhus, Astrakhan spotted fever, Kenya tick typhus, Indian tick typhus, or other names that designate the locality of occurrence while having distinct clinical features. It is a member of the spotted fever group and the most geographically dispersed species in the group, recognized in most of the regions bordering on the Mediterranean Sea and Black Sea, Israel, Kenya, and other parts of North, Central, and South Africa, and India. The prevailing vector is the brown dog tick, Rhipicephalus sanguineus. The bacterium was isolated by Emile Brumpt in 1932 and named after A. Conor, who in collaboration with A. Bruch, provided the first description of boutonneuse fever in Tunisia in 1910.

Mimivirus-dependent virus Sputnik is a subviral agent that reproduces in amoeba cells that are already infected by a certain helper virus; Sputnik uses the helper virus's machinery for reproduction and inhibits replication of the helper virus. It is known as a virophage, in analogy to the term bacteriophage.

Mimiviridae is a family of viruses. Amoeba and other protists serve as natural hosts. The family is divided in up to 4 subfamilies. Viruses in this family belong to the nucleocytoplasmic large DNA virus clade (NCLDV), also referred to as giant viruses.
Mamavirus is a large and complex virus in the Group I family Mimiviridae. The virus is exceptionally large, and larger than many bacteria. Mamavirus and other mimiviridae belong to nucleocytoplasmic large DNA virus (NCLDVs) family. Mamavirus can be compared to the similar complex virus mimivirus; mamavirus was so named because it is similar to but larger than mimivirus.
Marseillevirus is a genus of viruses, in the family Marseilleviridae. There are two species in this genus. It is the prototype of a family of nucleocytoplasmic large DNA viruses (NCLDV) of eukaryotes. It was isolated from amoeba.
A giant virus, sometimes referred to as a girus, is a very large virus, some of which are larger than typical bacteria. All known giant viruses belong to the phylum Nucleocytoviricota.

Megavirus is a viral genus containing a single identified species named Megavirus chilensis, phylogenetically related to Acanthamoeba polyphaga Mimivirus (APMV). In colloquial speech, Megavirus chilensis is more commonly referred to as just “Megavirus”. Until the discovery of pandoraviruses in 2013, it had the largest capsid diameter of all known viruses, as well as the largest and most complex genome among all known viruses.
A transpoviron is a plasmid-like genetic element found in the genomes of giant DNA viruses.
Afipia birgiae is a species in the Afipia bacterial genus. It is a gram-negative, oxidase-positive rod in the alpha-2 subgroup of the class Proteobacteria. It is motile by means of a single flagellum. Its type strain is 34632T.
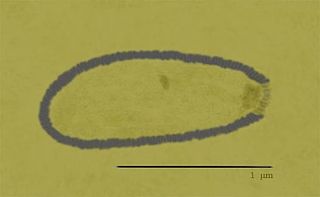
Pithovirus, first described in a 2014 paper, is a genus of giant virus known from two species, Pithovirus sibericum, which infects amoebas and Pithovirus massiliensis. It is a double-stranded DNA virus, and is a member of the nucleocytoplasmic large DNA viruses clade. The 2014 discovery was made when a viable specimen was found in a 30,000-year-old ice core harvested from permafrost in Siberia, Russia.

Mimivirus-dependent virus Zamilon, or Zamilon, is a virophage, a group of small DNA viruses that infect protists and require a helper virus to replicate; they are a type of satellite virus. Discovered in 2013 in Tunisia, infecting Acanthamoeba polyphaga amoebae, Zamilon most closely resembles Sputnik, the first virophage to be discovered. The name is Arabic for "the neighbour". Its spherical particle is 50–60 nm in diameter, and contains a circular double-stranded DNA genome of around 17 kb, which is predicted to encode 20 polypeptides. A related strain, Zamilon 2, has been identified in North America.

Faustovirus is a genus of giant virus which infects amoebae associated with humans. The virus was first isolated in 2015 and shown to be around 0.2 micrometers in diameter with a double stranded DNA genome of 466 kilobases predicted to encode 451 proteins. Although classified as a nucleocytoplasmic large DNA virus (NCLDV), faustoviruses share less than a quarter of their genes with other NCLDVs; however, ~46% are homologous to bacterial genes and the remainder are orphan genes (ORFans). Specifically, the gene encoding the major capsid protein (MCP) of faustovirus is different than that of its most closely related giant virus, asfivirus, as well as other NCLDVs. In asfivirus, the gene encoding MCP is a single genomic fragment of ~2000 base pairs (bp), however, in faustovirus the MCP is encoded by 13 exons separated by 12 large introns. The exons have a mean length of 149 bp and the introns have a mean length of 1,273 bp. The presence of introns in faustovirus genes is highly unusual for viruses.

Tupanvirus is a genus of viruses first described in 2018. The genus is composed of two species of virus that are in the giant virus group. Researchers discovered the first isolate in 2012 from deep water sediment samples taken at 3000m depth off the coast of Brazil. The second isolate was collected from a soda lake in Southern Nhecolândia, Brazil in 2014. They are named after Tupã (Tupan), a Guaraní thunder god, and the places they were found. These are the first viruses reported to possess genes for amino-acyl tRNA synthetases for all 20 standard amino acids.
Nucleocytoviricota is a phylum of viruses. Members of the phylum are also known as the nucleocytoplasmic large DNA viruses (NCLDV), which serves as the basis of the name of the phylum with the suffix -viricota for virus phylum. These viruses are referred to as nucleocytoplasmic because they are often able to replicate in both the host's cell nucleus and cytoplasm.