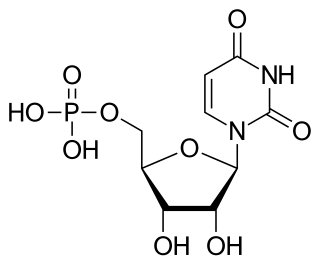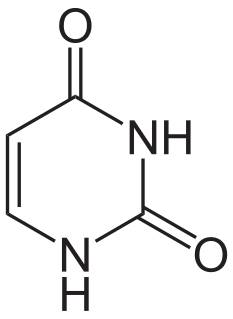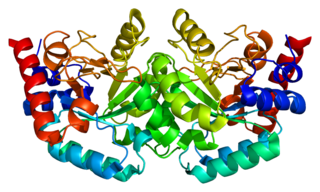
Uridine monophosphate synthase (UMPS) is the enzyme that catalyses the formation of uridine monophosphate (UMP), an energy-carrying molecule in many important biosynthetic pathways. In humans, the gene that codes for this enzyme is located on the long arm of chromosome 3 (3q13).
A salvage pathway is a pathway in which nucleotides are synthesized from intermediates in the degradative pathway for nucleotides.
Hypoxanthine-guanine phosphoribosyltransferase (HGPRT) is an enzyme encoded in humans by the HPRT1 gene.
DNA glycosylases are a family of enzymes involved in base excision repair, classified under EC number EC 3.2.2. Base excision repair is the mechanism by which damaged bases in DNA are removed and replaced. DNA glycosylases catalyze the first step of this process. They remove the damaged nitrogenous base while leaving the sugar-phosphate backbone intact, creating an apurinic/apyrimidinic site, commonly referred to as an AP site. This is accomplished by flipping the damaged base out of the double helix followed by cleavage of the N-glycosidic bond.
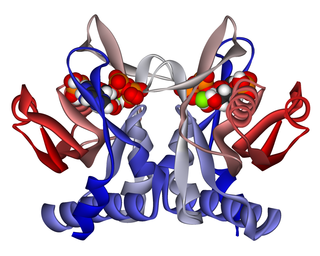
Adenine phosphoribosyltransferase (APRTase) is an enzyme encoded by the APRT gene, found in humans on chromosome 16. It is part of the Type I PRTase family and is involved in the nucleotide salvage pathway, which provides an alternative to nucleotide biosynthesis de novo in humans and most other animals. In parasitic protozoa such as giardia, APRTase provides the sole mechanism by which adenine can be produced. APRTase deficiency contributes to the formation of kidney stones (urolithiasis) and to potential kidney failure.
Pentosyltransferases are a type of glycosyltransferase that catalyze the transfer of a pentose.
Lumazine synthase (EC 2.5.1.78, 6,7-dimethyl-8-ribityllumazine synthase, 6,7-dimethyl-8-ribityllumazine synthase 2, 6,7-dimethyl-8-ribityllumazine synthase 1, lumazine synthase 2, lumazine synthase 1, type I lumazine synthase, type II lumazine synthase, RIB4, MJ0303, RibH, Pbls, MbtLS, RibH1 protein, RibH2 protein, RibH1, RibH2) is an enzyme with systematic name 5-amino-6-(D-ribitylamino)uracil butanedionetransferase. This enzyme catalyses the following chemical reaction

Nucleic acid metabolism is the process by which nucleic acids are synthesized and degraded. Nucleic acids are polymers of nucleotides. Nucleotide synthesis is an anabolic mechanism generally involving the chemical reaction of phosphate, pentose sugar, and a nitrogenous base. Destruction of nucleic acid is a catabolic reaction. Additionally, parts of the nucleotides or nucleobases can be salvaged to recreate new nucleotides. Both synthesis and degradation reactions require enzymes to facilitate the event. Defects or deficiencies in these enzymes can lead to a variety of diseases.
Pyrimidine biosynthesis occurs both in the body and through organic synthesis.
Purine metabolism refers to the metabolic pathways to synthesize and break down purines that are present in many organisms.

Rho-independent termination is a mechanism in prokaryotes that causes RNA transcription to stop and release the newly made RNA. In this mechanism, the mRNA contains a sequence that can base pair with itself to form a stem-loop structure 7–20 base pairs in length that is also rich in cytosine-guanine base pairs. C-G base pairs have significant base-stacking interactions and can form three hydrogen bonds between each other, resulting in a stable RNA duplex. Following the stem-loop structure is a chain of uracil residues. The bonds between uracil and adenine are very weak. A protein bound to RNA polymerase (nusA) binds to the stem-loop structure tightly enough to cause the polymerase to temporarily stall. This pausing of the polymerase coincides with transcription of the poly-uracil sequence. The weak Adenine-Uracil bonds lower the energy of destabilization for the RNA-DNA duplex, allowing it to unwind and dissociate from the RNA polymerase.

In enzymology, an ATP phosphoribosyltransferase is an enzyme that catalyzes the chemical reaction
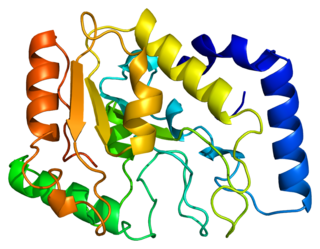
Uracil-DNA glycosylase, also known as UNG or UDG, is an enzyme. The human gene is well researched and orthologs exist ubiquitously among prokaryotes and eukaryotes and even in some DNA viruses. The first uracil DNA-glycosylase was isolated from Escherichia coli.

Cyclin-O is a protein that in humans is encoded by the CCNO gene.

Single-strand selective monofunctional uracil DNA glycosylase is an enzyme that in humans is encoded by the SMUG1 gene. SMUG1 is a glycosylase that removes uracil from single- and double-stranded DNA in nuclear chromatin, thus contributing to base excision repair.

Uridine-cytidine kinase-like 1 is an enzyme that in humans is encoded by the UCKL1 gene.
A phosphoribosyltransferase is a type of transferase enzyme.

Tegafur is a chemotherapeutic prodrug of 5-fluorouracil (5-FU) used in the treatment of cancers. It is a component of the combination drug tegafur/uracil. When metabolised, it becomes 5-FU.
The Nucleobase:Cation Symporter-2(NCS2) Family, also called the Nucleobase/Ascorbate Transporter(NAT) Family, consists of over 1000 sequenced proteins derived from gram-negative and gram-positive bacteria, archaea, fungi, plants and animals. The NCS2/NAT family is a member of the APC Superfamily of secondary carriers. Of the five known families of transporters that act on nucleobases, NCS2/NAT is the only one that is most widespread. Many functionally characterized members are specific for nucleobases including both purines and pyrimidines, but others are purine-specific. However, two closely related rat/human members of the family, SVCT1 and SVCT2, localized to different tissues of the body, co-transport L-ascorbate (vitamin C) and Na+ with a high degree of specificity and high affinity for the vitamin. Clustering of NCS2/NAT family members on the phylogenetic tree is complex, with bacterial proteins and eukaryotic proteins each falling into at least three distinct clusters. The plant and animal proteins cluster loosely together, but the fungal proteins branch from one of the three bacterial clusters forming a tighter grouping. E. coli possesses four distantly related paralogous members of the NCS2 family.