In molecular biology, a transcription factor (TF) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are 1500-1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome.

Histone acetyltransferases (HATs) are enzymes that acetylate conserved lysine amino acids on histone proteins by transferring an acetyl group from acetyl-CoA to form ε-N-acetyllysine. DNA is wrapped around histones, and, by transferring an acetyl group to the histones, genes can be turned on and off. In general, histone acetylation increases gene expression.

Histone deacetylases (EC 3.5.1.98, HDAC) are a class of enzymes that remove acetyl groups (O=C-CH3) from an ε-N-acetyl lysine amino acid on both histone and non-histone proteins. HDACs allow histones to wrap the DNA more tightly. This is important because DNA is wrapped around histones, and DNA expression is regulated by acetylation and de-acetylation. HDAC's action is opposite to that of histone acetyltransferase. HDAC proteins are now also called lysine deacetylases (KDAC), to describe their function rather than their target, which also includes non-histone proteins. In general, they suppress gene expression.

Histone acetyltransferase p300 also known as p300 HAT or E1A-associated protein p300 also known as EP300 or p300 is an enzyme that, in humans, is encoded by the EP300 gene. It functions as histone acetyltransferase that regulates transcription of genes via chromatin remodeling by allowing histone proteins to wrap DNA less tightly. This enzyme plays an essential role in regulating cell growth and division, prompting cells to mature and assume specialized functions (differentiate), and preventing the growth of cancerous tumors. The p300 protein appears to be critical for normal development before and after birth.
The p300-CBP coactivator family in humans is composed of two closely related transcriptional co-activating proteins :
- p300
- CBP
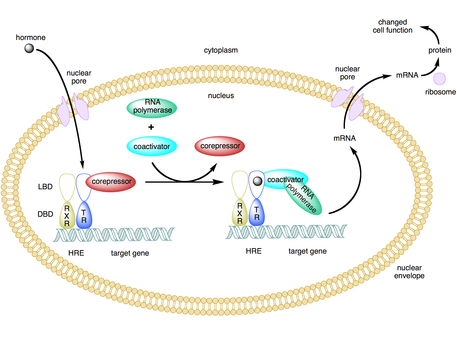
In molecular biology and genetics, transcription coregulators are proteins that interact with transcription factors to either activate or repress the transcription of specific genes. Transcription coregulators that activate gene transcription are referred to as coactivators while those that repress are known as corepressors. The mechanism of action of transcription coregulators is to modify chromatin structure and thereby make the associated DNA more or less accessible to transcription. In humans several dozen to several hundred coregulators are known, depending on the level of confidence with which the characterisation of a protein as a coregulator can be made. One class of transcription coregulators modifies chromatin structure through covalent modification of histones. A second ATP dependent class modifies the conformation of chromatin.

P300/CBP-associated factor (PCAF), also known as K(lysine) acetyltransferase 2B (KAT2B), is a human gene and transcriptional coactivator associated with p53.

CREB-binding protein, also known as CREBBP or CBP or KAT3A, is a coactivator encoded by the CREBBP gene in humans, located on chromosome 16p13.3. CBP has intrinsic acetyltransferase functions; it is able to add acetyl groups to both transcription factors as well as histone lysines, the latter of which has been shown to alter chromatin structure making genes more accessible for transcription. This relatively unique acetyltransferase activity is also seen in another transcription enzyme, EP300 (p300). Together, they are known as the p300-CBP coactivator family and are known to associate with more than 16,000 genes in humans; however, while these proteins share many structural features, emerging evidence suggests that these two co-activators may promote transcription of genes with different biological functions.

Histone acetylation and deacetylation are the processes by which the lysine residues within the N-terminal tail protruding from the histone core of the nucleosome are acetylated and deacetylated as part of gene regulation.
The nuclear receptor coactivator 1 (NCOA1), also called steroid receptor coactivator-1 (SRC-1), is a transcriptional coregulatory protein that contains several nuclear receptor–interacting domains and possesses intrinsic histone acetyltransferase activity. It is encoded by the gene NCOA1.

The nuclear receptor coactivator 3 also known as NCOA3 is a protein that, in humans, is encoded by the NCOA3 gene. NCOA3 is also frequently called 'amplified in breast 1' (AIB1), steroid receptor coactivator-3 (SRC-3), or thyroid hormone receptor activator molecule 1 (TRAM-1).

Histone acetyltransferase KAT2A is an enzyme that in humans is encoded by the KAT2A gene.
Histone acetyltransferase KAT5 is an enzyme that in humans is encoded by the KAT5 gene. It is also commonly identified as TIP60.

Nuclear receptor coactivator 6 is a protein that in humans is encoded by the NCOA6 gene.

K(lysine) acetyltransferase 6A (KAT6A), is an enzyme that, in humans, is encoded by the KAT6A gene. This gene is located on human chromosome 8, band 8p11.21.

TAF5-like RNA polymerase II p300/CBP-associated factor-associated factor 65 kDa subunit 5L is an enzyme that in humans is encoded by the TAF5L gene.
Nuclear receptor coregulators are a class of transcription coregulators that have been shown to be involved in any aspect of signaling by any member of the nuclear receptor superfamily. A comprehensive database of coregulators for nuclear receptors and other transcription factors was previously maintained at the Nuclear Receptor Signaling Atlas website which has since been replaced by the Signaling Pathways Project website.

In biochemistry, the KIX domain (kinase-inducible domain (KID) interacting domain) or CREB binding domain is a protein domain of the eukaryotic transcriptional coactivators CBP and P300. It serves as a docking site for the formation of heterodimers between the coactivator and specific transcription factors. Structurally, the KIX domain is a globular domain consisting of three α-helices and two short 310-helices.
While the cellular and molecular mechanisms of learning and memory have long been a central focus of neuroscience, it is only in recent years that attention has turned to the epigenetic mechanisms behind the dynamic changes in gene transcription responsible for memory formation and maintenance. Epigenetic gene regulation often involves the physical marking of DNA or associated proteins to cause or allow long-lasting changes in gene activity. Epigenetic mechanisms such as DNA methylation and histone modifications have been shown to play an important role in learning and memory.
Pharmacoepigenetics is an emerging field that studies the underlying epigenetic marking patterns that lead to variation in an individual's response to medical treatment.